2N5M
 
 | | Unveiling the structural determinants of KIAA0323 binding preference for NEDD8 | | Descriptor: | Protein KHNYN | | Authors: | Santonico, E, Nepravishta, R, Mattioni, A, Valentini, E, Mandaliti, W, Procopio, R, Iannuccelli, M, Castagnoli, L, Polo, S, Paci, M, Cesareni, G. | | Deposit date: | 2015-07-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unveiling the structural determinants of KIAA0323 binding preference for NEDD8.
To be Published
|
|
5ZBG
 
 | |
5V9J
 
 | | Crystal structure of catalytic domain of GLP with MS0105 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Dong, A, Zeng, H, Liu, J, Xiong, Y, Babault, N, Jin, J, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Wu, H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-23 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of catalytic domain of GLP with MS0105
to be published
|
|
5VCI
 
 | | RNA hairpin structure containing tetraloop/receptor motif, complexed with 2-MeImpG analogue | | Descriptor: | 5'-O-[(S)-hydroxy(4-methyl-1H-imidazol-5-yl)phosphoryl]guanosine, RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*CP*CP*CP*GP*AP*AP*AP*GP*G)-3'), RNA (5'-R(P*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2017-03-31 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Rationale for the Enhanced Catalysis of Nonenzymatic RNA Primer Extension by a Downstream Oligonucleotide.
J. Am. Chem. Soc., 140, 2018
|
|
5VAP
 
 | | Crystal structure of eVP30 C-terminus and eNP peptide | | Descriptor: | Minor nucleoprotein VP30, NP | | Authors: | XU, W, WU, C, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ebola virus VP30 and nucleoprotein interactions modulate viral RNA synthesis.
Nat Commun, 8, 2017
|
|
2B8U
 
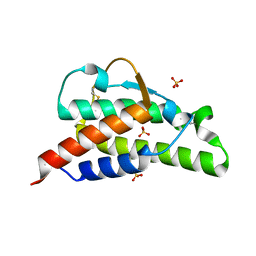 | | Crystal structure of wildtype human Interleukin-4 | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
3ZXJ
 
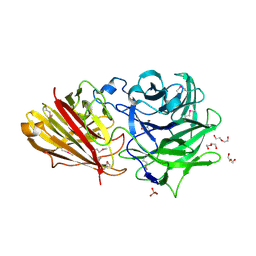 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HIAXHD3, ... | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2B8Y
 
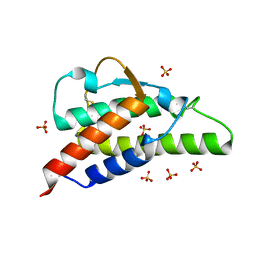 | | Crystal structure of the interleukin-4 variant T13DF82D | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B91
 
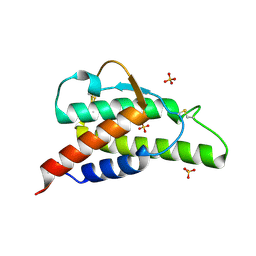 | | Crystal structure of the interleukin-4 variant F82DR85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
3ZXK
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIAXHD3, alpha-L-arabinofuranose-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5VCN
 
 | | THE CRYSTAL STRUCTURE OF DER P 1 ALLERGEN COMPLEXED WITH FAB FRAGMENT OF MAB 5H8 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Osinski, T, Majorek, K.A, Pomes, A, Offermann, L.R, Osinski, S, Glesner, J, Vailes, L.D, Chapman, M.D, Minor, W, Chruszcz, M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Der p 1-Antibody Complexes and Comparison with Complexes of Proteins or Peptides with Monoclonal Antibodies.
J. Immunol., 195, 2015
|
|
2B8Z
 
 | | Crystal structure of the interleukin-4 variant R85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
4UIB
 
 | | Crystal structure of 3p in complex with tafCPB | | Descriptor: | (2S)-6-AMINO-2-[[(1R)-1-(CYCLOHEXYLMETHYL)-2-OXO-2-[[(2S)-1,7,7-TRIMETHYLNORBORNAN-2-YL]AMINO]ETHYL]ARBAMOYLAMINO]HEXANOIC ACID, ACETATE ION, CARBOXYPEPTIDASE B, ... | | Authors: | Halland, N, Broenstrup, M, Czech, J, Czechtizky, W, Evers, A, Follmann, M, Kohlmann, M, Schiell, M, Kurz, M, Schreuder, H.A, Kallus, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (Tafia) from Natural Product Anabaenopeptin.
J.Med.Chem., 58, 2015
|
|
3IKI
 
 | | 5-SMe-dU containing DNA octamer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(US2)P*AP*CP*AP*C)-3', MAGNESIUM ION | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | Deposit date: | 2009-08-05 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Hydrogen bond formation between the naturally modified nucleobase and phosphate backbone.
Nucleic Acids Res., 40, 2012
|
|
4UIA
 
 | | Crystal structure of 3a in complex with tafCPB | | Descriptor: | (2S)-6-azanyl-2-[[(2R)-3-cyclohexyl-1-(3-methylbutylamino)-1-oxidanylidene-propan-2-yl]carbamoylamino]hexanoic acid, CARBOXYPEPTIDASE B, ZINC ION | | Authors: | Halland, N, Broenstrup, M, Czech, J, Czechtizky, W, Evers, A, Follmann, M, Kohlmann, M, Schiell, M, Kurz, M, Schreuder, H.A, Kallus, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Novel Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (Tafia) from Natural Product Anabaenopeptin.
J.Med.Chem., 58, 2015
|
|
5ZOF
 
 | | Crystal Structure of D181A/R192F hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), DNA (5'-D(*GP*CP*CP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
2BCO
 
 | | X-ray structure of succinylglutamate desuccinalase from Vibrio Parahaemolyticus (RIMD 2210633) at the resolution 2.3 A, Northeast Structural Genomics Target Vpr14 | | Descriptor: | Succinylglutamate desuccinylase, ZINC ION | | Authors: | Kuzin, A.P, Abashidze, M, Forouhar, F, Benach, J, Zhou, W, Acton, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-19 | | Release date: | 2005-10-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | X-ray structure of succinylglutamate desuccinalase from Vibrio Parahaemolyticus (RIMD 2210633) at the resolution 2.3 A, Northeast Structural Genomics Target Vpr14
To be Published
|
|
5UNH
 
 | | Synchrotron structure of human angiotensin II type 2 receptor in complex with compound 2 (N-[(furan-2-yl)methyl]-N-(4-oxo-2-propyl-3-{[2'-(2H-tetrazol-5-yl)[1,1'- biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)benzamide) | | Descriptor: | N-[(furan-2-yl)methyl]-N-(4-oxo-2-propyl-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)benzamide, Soluble cytochrome b562,Type-2 angiotensin II receptor | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
2ARG
 
 | | FORMATION OF AN AMINO ACID BINDING POCKET THROUGH ADAPTIVE ZIPPERING-UP OF A LARGE DNA HAIRPIN LOOP, NMR, 9 STRUCTURES | | Descriptor: | ARGININEAMIDE, DNA APTAMER [5'-D (*TP*GP*AP*CP*CP*AP*GP*GP*GP*CP*AP*AP*AP*CP*GP*GP*TP*AP* GP*GP*TP*GP*AP*GP*TP*GP*GP*TP*CP*A)-3'] | | Authors: | Lin, C.H, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1998-08-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Formation of an amino-acid-binding pocket through adaptive zippering-up of a large DNA hairpin loop.
Chem.Biol., 5, 1998
|
|
2GPM
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), RNA (5'-R(*(0C)P*(0U)P*(0G)P*(0G)P*(0G)P*(0C)P*(0G)P*(0G))-3') | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-18 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5ZHN
 
 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-({4-[(octylamino)methyl]phenyl}methyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
3ILF
 
 | | Crystal structure of porphyranase A (PorA) in complex with neo-porphyrotetraose | | Descriptor: | 6-O-sulfo-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-6-O-sulfo-alpha-L-galactopyranose-(1-3)-alpha-D-galactopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Hehemann, J.H, Correc, G, Barbeyron, T, Helbert, W, Michel, G, Czjzek, M. | | Deposit date: | 2009-08-07 | | Release date: | 2010-04-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota.
Nature, 464, 2010
|
|
2GTA
 
 | | Crystal Structure of the putative pyrophosphatase YPJD from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR428. | | Descriptor: | Hypothetical protein ypjD, SODIUM ION | | Authors: | Vorobiev, S.M, Zhou, W, Seetharaman, J, Wang, D, Ma, L.C, Acton, T, Xio, R, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-27 | | Release date: | 2006-05-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the putative pyrophosphatase YPJD from Bacillus subtilis.
To be Published
|
|
2AZO
 
 | | DNA MISMATCH REPAIR PROTEIN MUTH FROM E. COLI | | Descriptor: | MUTH | | Authors: | Yang, W. | | Deposit date: | 1997-11-20 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases.
EMBO J., 17, 1998
|
|
5VER
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Z | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
