1JV0
 
 | | THE CRYSTAL STRUCTURE OF THE ZINC(II) ADDUCT OF THE CAI MICHIGAN 1 VARIANT | | Descriptor: | 1,2-ETHANEDIOL, CARBONIC ANHYDRASE I, CHLORIDE ION, ... | | Authors: | Briganti, F, Ferraroni, M, Chegwidden, W.R, Scozzafava, A, Supuran, C.T, Tilli, S. | | Deposit date: | 2001-08-28 | | Release date: | 2001-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a zinc-activated variant of human carbonic anhydrase I, CA I Michigan 1: evidence for a second zinc binding site involving arginine coordination
Biochemistry, 41, 2002
|
|
3BBC
 
 | |
3BGX
 
 | | E. coli Thymidylate Synthase C146S mutant complexed with dTMP and MTF | | Descriptor: | N-({4-[(6aR)-3-amino-1-oxo-1,2,5,6,6a,7-hexahydroimidazo[1,5-f]pteridin-8(9H)-yl]phenyl}carbonyl)-L-glutamic acid, SULFATE ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Roberts, S.A, Montfort, W.R. | | Deposit date: | 2007-11-27 | | Release date: | 2008-12-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Studies of Early Events in Catalysis by Thymidylate Synthase
To be Published
|
|
1L1E
 
 | | Crystal Structure of Mycolic Acid Cyclopropane Synthase PcaA Complexed with S-adenosyl-L-homocysteine | | Descriptor: | CARBONATE ION, S-ADENOSYL-L-HOMOCYSTEINE, mycolic acid synthase | | Authors: | Huang, C.-C, Smith, C.V, Glickman, M.S, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-02-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of mycolic acid cyclopropane synthases from Mycobacterium tuberculosis.
J.Biol.Chem., 277, 2002
|
|
1HKV
 
 | | mycobacterium diaminopimelate dicarboxylase (lysa) | | Descriptor: | DIAMINOPIMELATE DECARBOXYLASE, LYSINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gokulan, K, Rupp, B, Pavelka Jr, M.S, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-03-11 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Mycobacterium Tuberculosis Diaminopimelate Decarboxylase, an Essential Enzyme in Bacterial Lysine Biosynthesis
J.Biol.Chem., 278, 2003
|
|
3QXE
 
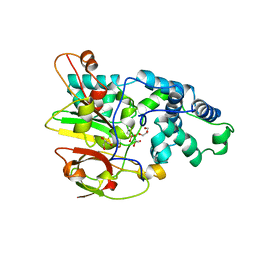 | | Crystal Structure of Co-type Nitrile Hydratase from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
3QQX
 
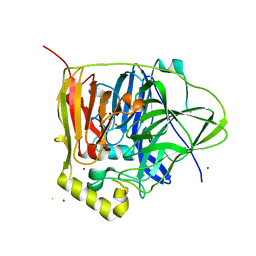 | | Reduced Native Intermediate of the Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (I) ION, COPPER (II) ION, ... | | Authors: | Montfort, W.R, Roberts, S.A, Singh, S.K. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CueO E506D Mutant: Crystal Structure of Reduced Native Intermediate, Kinetics, and Impairment of Product Release
To be Published
|
|
3TYG
 
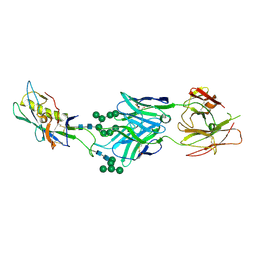 | | Crystal structure of broad and potent HIV-1 neutralizing antibody PGT128 in complex with a glycosylated engineered gp120 outer domain with miniV3 (eODmV3) | | Descriptor: | Envelope glycoprotein gp160, PGT128 heavy chain, Ig gamma-1 chain C region, ... | | Authors: | Pejchal, R, Huang, P.S, Schief, W.R, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2011-09-25 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield.
Science, 334, 2011
|
|
3C77
 
 | |
3U8P
 
 | | Cytochrome b562 integral fusion with EGFP | | Descriptor: | Cytochrome b562 integral fusion with enhanced green fluorescent protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arpino, J, Czapinska, H, Piasecka, A, Edwards, W.R, Barker, P, Gajda, M, Bochtler, M, Jones, D.D. | | Deposit date: | 2011-10-17 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for efficient chromophore communication and energy transfer in a constructed didomain protein scaffold.
J.Am.Chem.Soc., 134, 2012
|
|
3TJ8
 
 | | Crystal structure of Helicobacter pylori UreE bound to Ni2+ | | Descriptor: | FORMIC ACID, NICKEL (II) ION, Urease accessory protein ureE | | Authors: | Banaszak, K, Bellucci, M, Zambelli, B, Rypniewski, W.R, Ciurli, S. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Crystallographic and X-ray absorption spectroscopic characterization of Helicobacter pylori UreE bound to Ni2+ and Zn2+ reveals a role for the disordered C-terminal arm in metal trafficking.
Biochem.J., 441, 2012
|
|
3RKR
 
 | | Crystal structure of a metagenomic short-chain oxidoreductase (SDR) in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, Short chain oxidoreductase | | Authors: | Mayerhofer, H, Bijtenhoorn, P, Streit, W.R, Mueller-Dieckmann, J. | | Deposit date: | 2011-04-18 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A novel metagenomic short-chain dehydrogenase/reductase attenuates Pseudomonas aeruginosa biofilm formation and virulence on Caenorhabditis elegans.
Plos One, 6, 2011
|
|
1LIA
 
 | |
3BXV
 
 | |
3C76
 
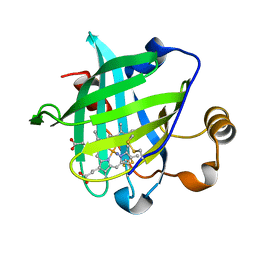 | |
3UBP
 
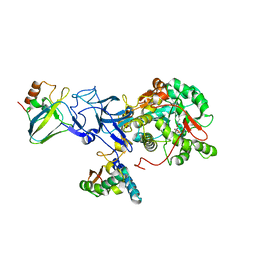 | | DIAMIDOPHOSPHATE INHIBITED BACILLUS PASTEURII UREASE | | Descriptor: | DIAMIDOPHOSPHATE, NICKEL (II) ION, PROTEIN (UREASE ALPHA SUBUNIT), ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Miletti, S, Mangani, S, Ciurli, S. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels.
Structure Fold.Des., 7, 1999
|
|
1NHW
 
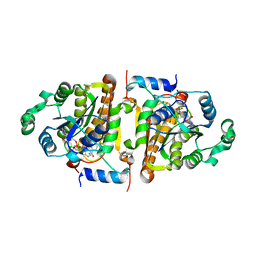 | | Crystal Structure Analysis of Plasmodium falciparum enoyl-acyl-carrier-protein reductase | | Descriptor: | 2-(2,4-DICHLORO-PHENYLAMINO)-PHENOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, enoyl-acyl carrier reductase | | Authors: | Perozzo, R, Kuo, M, Sidhu, A.S, Valiyaveettil, J.T, Bittman, R, Jacobs Jr, W.R, Fidock, D.A, Sacchettini, J.C. | | Deposit date: | 2002-12-19 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Elucidation of the Specificity of the Antibacterial Agent Triclosan for
Malarial Enoyl Acyl Carrier Protein Reductase Year
J.Biol.Chem., 277, 2002
|
|
1NP4
 
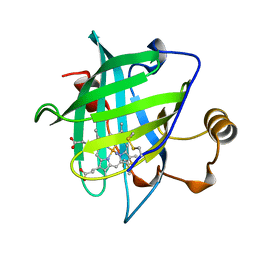 | | CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS | | Descriptor: | AMMONIA, PROTEIN (NITROPHORIN 4), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Andersen, J.F, Weichsel, A, Champagne, D.E, Balfour, C.A, Montfort, W.R. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of nitrophorin 4 at 1.5 A resolution: transport of nitric oxide by a lipocalin-based heme protein.
Structure, 6, 1998
|
|
1NP1
 
 | |
1NNU
 
 | | Crystal Structure Analysis of Plasmodium falciparum enoyl-acyl-carrier-protein reductase with Triclosan Analog | | Descriptor: | 6-(4-CHLORO-2-HYDROXY-PHENOXY)-NAPHTHALEN-2-OL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, enoyl-acyl carrier reductase | | Authors: | Perozzo, R, Kuo, M, Sidhu, A.S, Valiyaveettil, J.T, Bittman, R, Jacobs Jr, W.R, Fidock, D.A, Sacchettini, J.C. | | Deposit date: | 2003-01-14 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Elucidation of the Specificity of the Antibacterial Agent Triclosan for
Malarial Enoyl Acyl Carrier Protein Reductase
J.Biol.Chem., 277, 2002
|
|
1NP2
 
 | |
1P45
 
 | | Targeting tuberculosis and malaria through inhibition of enoyl reductase: compound activity and structural data | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Kuo, M.R, Morbidoni, H.R, Alland, D, Sneddon, S.F, Gourlie, B.B, Staveski, M.M, Leonard, M, Gregory, J.S, Janjigian, A.D, Yee, C, Musser, J.M, Kreiswirth, B.N, Iwamoto, H, Perozzo, R, Jacobs Jr, W.R, Sacchettini, J.C, Fidock, D.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-21 | | Release date: | 2003-09-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Targeting tuberculosis and malaria through inhibition of Enoyl reductase: compound activity and structural data.
J.Biol.Chem., 278, 2003
|
|
1P52
 
 | | Structure of Arginine kinase E314D mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, D-ARGININE, ... | | Authors: | Pruett, P.S, Azzi, A, Clark, S.A, Yousef, M.S, Gattis, J.L, Somasundarum, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase.
J.Biol.Chem., 278, 2003
|
|
1NL3
 
 | | CRYSTAL STRUCTURE OF THE SECA PROTEIN TRANSLOCATION ATPASE FROM MYCOBACTERIUM TUBERCULOSIS in APO FORM | | Descriptor: | PREPROTEIN TRANSLOCASE SECA 1 SUBUNIT | | Authors: | Sharma, V, Arockiasamy, A, Ronning, D.R, Savva, C.G, Holzenburg, A, Braunstein, M, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-01-06 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of M. tuberculosis SecA, A Preprotein Translocating ATPase
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NCC
 
 | | CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IGG2A-KAPPA NC41 FAB (HEAVY CHAIN), ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
J.Mol.Biol., 227, 1992
|
|
