6DU0
 
 | |
6DI6
 
 | |
6DTV
 
 | |
6DLV
 
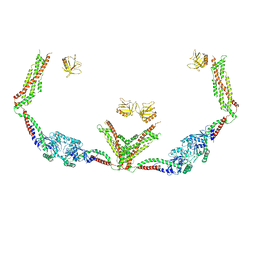 | | Cryo-EM of the GTP-bound human dynamin-1 polymer assembled on the membrane in the super constricted state | | Descriptor: | Dynamin-1 | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
6DTZ
 
 | | Crystal structure of eukaryotic DNA primase large subunit iron-sulfur cluster domain, Y397F mutant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, ... | | Authors: | Salay, L.E, Chazin, W.J. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Yeast require redox switching in DNA primase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7TL3
 
 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 5YF431 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, IRON/SULFUR CLUSTER | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.066 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
7TL4
 
 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 6YF | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, IRON/SULFUR CLUSTER | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
7TL2
 
 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 5YF412 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, GLYCEROL, ... | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
6E27
 
 | | The CARD9 CARD domain-swapped dimer with a zinc ion bound to one of the two zinc binding sites | | Descriptor: | Caspase recruitment domain-containing protein 9, ZINC ION | | Authors: | Holliday, M.J, Ferrao, R, Boenig, G, Deuber, E.C, Fairbrother, W.J. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Picomolar zinc binding modulates formation of Bcl10-nucleating assemblies of the caspase recruitment domain (CARD) of CARD9.
J. Biol. Chem., 293, 2018
|
|
6E28
 
 | | The CARD9 CARD domain-swapped dimer | | Descriptor: | Caspase recruitment domain-containing protein 9 | | Authors: | Holliday, M.J, Ferrao, R, Boenig, G, Deuber, E.C, Fairbrother, W.J. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Picomolar zinc binding modulates formation of Bcl10-nucleating assemblies of the caspase recruitment domain (CARD) of CARD9.
J. Biol. Chem., 293, 2018
|
|
1TW6
 
 | | Structure of an ML-IAP/XIAP chimera bound to a 9mer peptide derived from Smac | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Vucic, D, Wallweber, H.J.A, Das, K, Shin, H, Elliott, L.O, Kadkhodayan, S, Deshayes, K, Salvesen, G.S, Fairbrother, W.J. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.713 Å) | | Cite: | Engineering ML-IAP to produce an extraordinarily potent caspase 9 inhibitor: implications for Smac-dependent anti-apoptotic activity of ML-IAP
Biochem.J., 385, 2005
|
|
6TXS
 
 | | The structure of the FERM domain and helical linker of human moesin bound to a CD44 peptide | | Descriptor: | CD44 antigen, Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Kelly, J.J, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
6TXQ
 
 | | The high resolution structure of the FERM domain and helical linker of human moesin | | Descriptor: | ACETATE ION, Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Kelly, J.J, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
1TPT
 
 | | THREE-DIMENSIONAL STRUCTURE OF THYMIDINE PHOSPHORYLASE FROM ESCHERICHIA COLI AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | SULFATE ION, THYMIDINE PHOSPHORYLASE, THYMINE | | Authors: | Walter, M.R, Cook, W.J, Cole, L.B, Short, S.A, Koszalka, G.W, Krenitsky, T.A, Ealick, S.E. | | Deposit date: | 1990-06-14 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of thymidine phosphorylase from Escherichia coli at 2.8 A resolution.
J.Biol.Chem., 265, 1990
|
|
6E26
 
 | |
6E25
 
 | |
1UV6
 
 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with carbamylcholine | | Descriptor: | 2-[(AMINOCARBONYL)OXY]-N,N,N-TRIMETHYLETHANAMINIUM, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-01-15 | | Release date: | 2004-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
7UQA
 
 | | Crystal structure of the small Ultra-Red Fluorescent Protein (smURFP) | | Descriptor: | CHLORIDE ION, SODIUM ION, small Ultra-Red Fluorescent Protein (smURFP) | | Authors: | Maiti, A, Buffalo, C.Z, Saurabh, S, Montecinos-Franjola, F, Hachey, J.S, Conlon, W.J, Tran, G.N, Drobizhev, M, Moerner, W.E, Ghosh, P, Matsuo, H, Tsien, R.Y, Lin, J.Y, Rodriguez, E.A. | | Deposit date: | 2022-04-19 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural and photophysical characterization of the small ultra-red fluorescent protein.
Nat Commun, 14, 2023
|
|
1UX2
 
 | | X-ray structure of acetylcholine binding protein (AChBP) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-02-18 | | Release date: | 2004-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
8UCU
 
 | | Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-09-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6V6A
 
 | | Inhibitory scaffolding of the ancient MAPK, ERK7 | | Descriptor: | 1,2-ETHANEDIOL, Apical Cap Protein 9 (AC9), Mitogen-activated protein kinase | | Authors: | Dewangan, P.S, O'Shaughnessy, W.J, Back, P.S, Hu, X, Bradley, P.J, Reese, M.L. | | Deposit date: | 2019-12-04 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ancient MAPK ERK7 is regulated by an unusual inhibitory scaffold required forToxoplasmaapical complex biogenesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8V6I
 
 | | DNA elongation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (14.06 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6J
 
 | | DNA elongation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.11 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5O
 
 | | Tetramer core subcomplex (conformation 3) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.99 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5N
 
 | | Tetramer core subcomplex (conformation 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.56 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
