5F5O
 
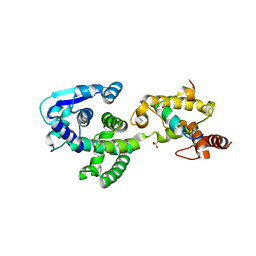 | | Crystal structure of Marburg virus nucleoprotein core domain bound to VP35 regulation peptide | | Descriptor: | Nucleoprotein, Peptide from Polymerase cofactor VP35, SULFATE ION | | Authors: | Guo, Y, Liu, B.C, Liu, X, Li, G.B, Wang, W.M, Dong, S.S, Wang, W.J. | | Deposit date: | 2015-12-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus
J. Virol., 91, 2017
|
|
5FJ9
 
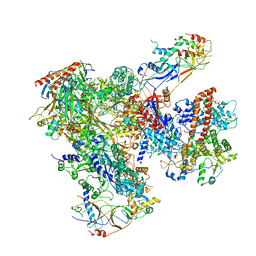 | | Cryo-EM structure of yeast apo RNA polymerase III at 4.6 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
5FJA
 
 | | Cryo-EM structure of yeast RNA polymerase III at 4.7 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
6CYI
 
 | | Grp94 N-domain bound to NEOCA | | Descriptor: | 5'-N-(2-HYDROXYL)ETHYL CARBOXYAMIDO ADENOSINE, Endoplasmin, TETRAETHYLENE GLYCOL, ... | | Authors: | Huck, J.D, Aw, W.J, Gewirth, D.T. | | Deposit date: | 2018-04-05 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7565825 Å) | | Cite: | NECA derivatives exploit the paralog-specific properties of the site 3 side pocket of Grp94, the endoplasmic reticulum Hsp90.
J.Biol.Chem., 294, 2019
|
|
5FJ8
 
 | | Cryo-EM structure of yeast RNA polymerase III elongation complex at 3. 9 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
3FAU
 
 | | Crystal Structure of human small-MutS related domain | | Descriptor: | NEDD4-binding protein 2 | | Authors: | Kim, T.G, Kwon, T.H, Ryu, E.K, Min, K, Heo, S.-D, Song, K.M, Jun, W.J, Jung, E. | | Deposit date: | 2008-11-18 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Strcutral Dynamincs of the Endonuclease Small-MutS Related Domains of BCL3 binding protein
To be Published
|
|
3EWV
 
 | | Crystal Structure of calmodulin complexed with a peptide | | Descriptor: | CALCIUM ION, Calmodulin, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Jiang, T, Cao, P, Gong, Y, Yu, H.J, Gui, W.J, Zhang, W.T. | | Deposit date: | 2008-10-16 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the mechanism of calmodulin binding to death receptors.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3EWT
 
 | | Crystal Structure of calmodulin complexed with a peptide | | Descriptor: | CALCIUM ION, Calmodulin, Tumor necrosis factor receptor superfamily member 6 | | Authors: | Jiang, T, Cao, P, Gong, Y, Yu, H.J, Gui, W.J, Zhang, W.T. | | Deposit date: | 2008-10-16 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the mechanism of calmodulin binding to death receptors.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5C1M
 
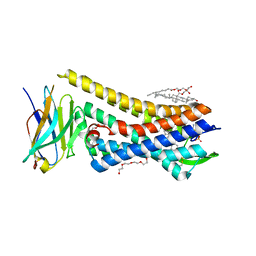 | | Crystal structure of active mu-opioid receptor bound to the agonist BU72 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3S,3aR,5aR,6R,11bR,11cS)-3a-methoxy-3,14-dimethyl-2-phenyl-2,3,3a,6,7,11c-hexahydro-1H-6,11b-(epiminoethano)-3,5a-methanonaphtho[2,1-g]indol-10-ol, CHOLESTEROL, ... | | Authors: | Huang, W.J, Manglik, A, Venkatakrishnan, A.J, Laeremans, T, Feinberg, E.N, Sanborn, A.L, Kato, H.E, Livingston, K.E, Thorsen, T.S, Kling, R, Granier, S, Gmeiner, P, Husbands, S.M, Traynor, J.R, Weis, W.I, Steyaert, J, Dror, R.O, Kobilka, B.K. | | Deposit date: | 2015-06-15 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insights into mu-opioid receptor activation.
Nature, 524, 2015
|
|
5D6J
 
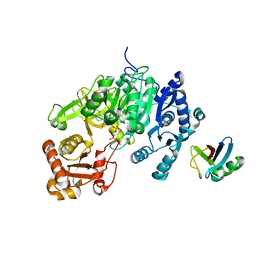 | | Crystal structure of a mycobacterial protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-CoA synthase, MAGNESIUM ION, ... | | Authors: | Li, W.J, Bi, L.J. | | Deposit date: | 2015-08-12 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of FadD32, an enzyme essential for mycolic acid biosynthesis in mycobacteria.
Sci Rep, 5, 2015
|
|
5D6N
 
 | | Crystal structure of a mycobacterial protein | | Descriptor: | Acyl-CoA synthase | | Authors: | Li, W.J, Bi, L.J. | | Deposit date: | 2015-08-12 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of FadD32, an enzyme essential for mycolic acid biosynthesis in mycobacteria.
Sci Rep, 5, 2015
|
|
8FAY
 
 | | Human MUTYH adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*CP*AP*(NR1)P*GP*TP*CP*T)-3'), ... | | Authors: | Trasvina-Arenas, C.H, Lin, W.J, Demir, M, Fisher, A.J, David, S.S, Horvath, M.P. | | Deposit date: | 2022-11-29 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Human MUTYH adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G)
To Be Published
|
|
1AHD
 
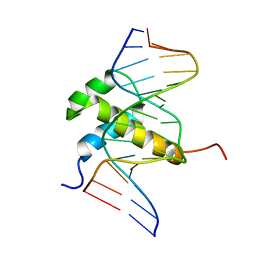 | | DETERMINATION OF THE NMR SOLUTION STRUCTURE OF AN ANTENNAPEDIA HOMEODOMAIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), Homeotic protein antennapedia | | Authors: | Billeter, M, Qian, Y.Q, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1993-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex.
J.Mol.Biol., 234, 1993
|
|
5Z80
 
 | | Solution structure for the 1:1 complex of a platinum(II)-based tripod bound to a hybrid-1 human telomeric G-quadruplex | | Descriptor: | 4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]-N,N-bis[4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]phenyl]aniline, G-quadruplex DNA (26-MER) | | Authors: | Liu, W.T, Zhong, Y.F, Liu, L.Y, Zeng, W.J, Wang, F.Y, Yang, D.Z, Mao, Z.W. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding
Nat Commun, 9, 2018
|
|
5XBN
 
 | |
5Z8F
 
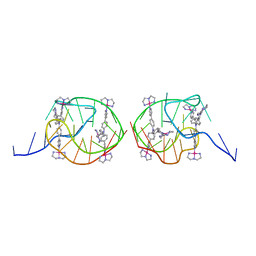 | | Solution structure for the unique dimeric 4:2 complex of a platinum(II)-based tripod bound to a hybrid-1 human telomeric G-quadruplex | | Descriptor: | 4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]-N,N-bis[4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]phenyl]aniline, G-quadruplex DNA (26-MER) | | Authors: | Liu, W.T, Zhong, Y.F, Liu, L.Y, Zeng, W.J, Wang, F.Y, Yang, D.Z, Mao, Z.W. | | Deposit date: | 2018-01-31 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding
Nat Commun, 9, 2018
|
|
8BG9
 
 | | Murine amyloid-beta filaments with the Arctic mutation (E22G) from APP(NL-G-F) mouse brains | ABeta | | Descriptor: | Amyloid-beta protein 40 | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordber, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
8BG0
 
 | | Amyloid-beta tetrameric filaments with the Arctic mutation (E22G) from Alzheimer's disease brains | ABeta40 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordber, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (1.99 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
8BFZ
 
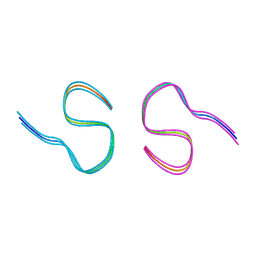 | | Amyloid-beta 42 filaments extracted from the human brain with Arctic mutation (E22G) of Alzheimer's disease | ABeta42 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordberg, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
1LXT
 
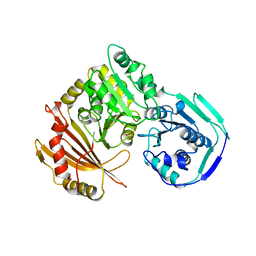 | | STRUCTURE OF PHOSPHOTRANSFERASE PHOSPHOGLUCOMUTASE FROM RABBIT | | Descriptor: | CADMIUM ION, PHOSPHOGLUCOMUTASE (DEPHOSPHO FORM), SULFATE ION | | Authors: | Ray Junior, W.J, Baranidharan, S, Liu, Y. | | Deposit date: | 1996-07-28 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of rabbit muscle phosphoglucomutase refined at 2.4 A resolution.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
3ZPN
 
 | | Structure of Psb28 | | Descriptor: | PHOTOSYSTEM II REACTION CENTER PSB28 PROTEIN | | Authors: | Bialek, W.J, Michoux, F, Nixon, P.J, Murray, J.W. | | Deposit date: | 2013-02-28 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Crystal Structure of the Psb28 Accessory Factor of Thermosynechococcus Elongatus Photosystem II at 2.3 A
Photosynth.Res., 117, 2013
|
|
7WI6
 
 | | Cryo-EM structure of LY341495/NAM-bound mGlu3 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
7WIH
 
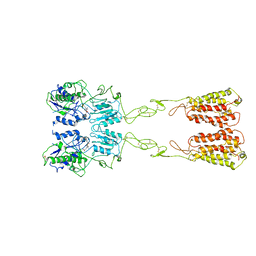 | | Cryo-EM structure of LY2794193-bound mGlu3 | | Descriptor: | (1S,2S,4S,5R,6S)-2-amino-4-[(3-methoxybenzene-1-carbonyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
7WI8
 
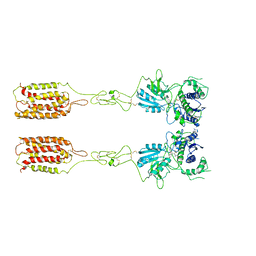 | | Cryo-EM structure of inactive mGlu3 bound to LY341495 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
7X44
 
 | |
