1GI5
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-5-METHOXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
6BMD
 
 | |
6BHH
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, SULFATE ION, ... | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6BHE
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6BIG
 
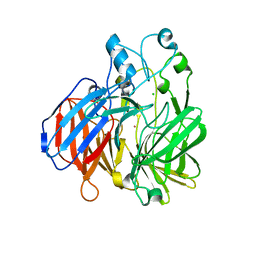 | | Crystal structure of cobalt-substituted Synechocystis ACO | | Descriptor: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, COBALT (II) ION | | Authors: | Sui, X, Shi, W, Kiser, P.D. | | Deposit date: | 2017-11-02 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Preparation and characterization of metal-substituted carotenoid cleavage oxygenases.
J. Biol. Inorg. Chem., 23, 2018
|
|
6BL2
 
 | | Novel Modes of Inhibition of Wild-Type IDH1: Direct Covalent Modification of His315 with Cmpd15 | | Descriptor: | 3-[(6aS,7S,9S,10aS)-9-cyano-7-methyl-8-oxo-2-(phenylamino)-6,6a,7,8,9,10-hexahydrobenzo[h]quinazolin-10a(5H)-yl]benzoic acid, CALCIUM ION, ISOCITRIC ACID, ... | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2017-11-09 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
1GQF
 
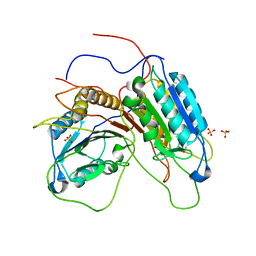 | |
6BSJ
 
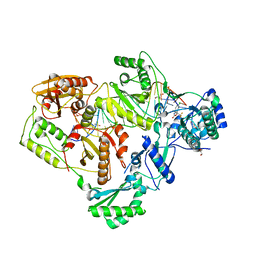 | | Structure of HIV-1 RT complexed with an RNA/DNA hybrid sequence non-preferred for RNA hydrolysis | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*AP*TP*GP*CP*CP*TP*AP*TP*AP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Tian, L, Kim, M, Yang, W. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BSI
 
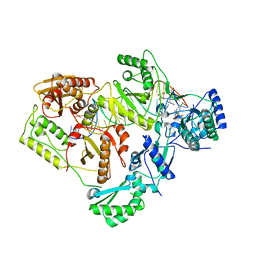 | | Structure of HIV-1 RT complexed with an RNA/DNA hybrid containing the polypurine-tract sequence | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*TP*TP*TP*TP*CP*TP*TP*TP*TP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Tian, L, Kim, M, Yang, W. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1H7U
 
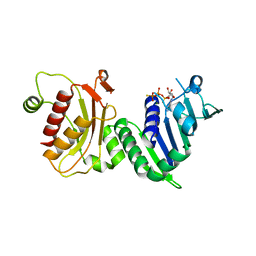 | | hPMS2-ATPgS | | Descriptor: | MAGNESIUM ION, MISMATCH REPAIR ENDONUCLEASE PMS2, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Guarne, A, Junop, M.S, Yang, W. | | Deposit date: | 2001-07-10 | | Release date: | 2001-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Function of the N-Terminal 40 kDa Fragment of Human Pms2: A Monomeric Ghl ATPase
Embo J., 20, 2001
|
|
6CQE
 
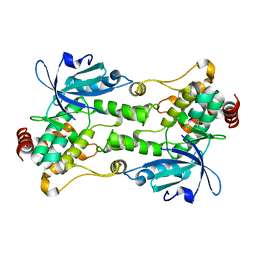 | | Crystal structure of HPK1 kinase domain S171A mutant | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Wu, P, Lehoux, I, Mortara, K, Franke, Y, Wang, W. | | Deposit date: | 2018-03-15 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.886 Å) | | Cite: | Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Structure, 27, 2019
|
|
6CV0
 
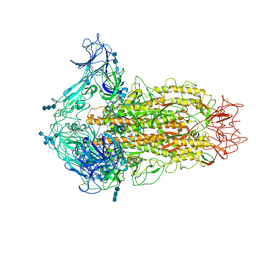 | | Cryo-electron microscopy structure of infectious bronchitis coronavirus spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Zheng, Y, Yang, Y, Liu, C, Geng, Q, Luo, C, Zhang, W, Li, F. | | Deposit date: | 2018-03-27 | | Release date: | 2018-04-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins.
PLoS Pathog., 14, 2018
|
|
6CTA
 
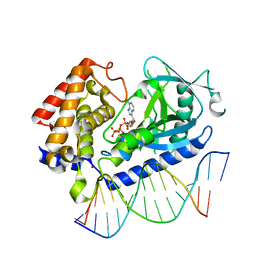 | | Structure of the human cGAS-DNA complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*GP*A)-3'), ... | | Authors: | Zhou, W, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-03-22 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.779 Å) | | Cite: | Structure of the Human cGAS-DNA Complex Reveals Enhanced Control of Immune Surveillance.
Cell, 174, 2018
|
|
1FUW
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF A DOUBLE MUTANT SINGLE-CHAIN MONELLIN(SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | MONELLIN | | Authors: | Sung, Y.H, Shin, J, Jung, J, Lee, W. | | Deposit date: | 2000-09-18 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics, and stability of a double mutant single-chain monellin. structural origin of sweetness.
J.Biol.Chem., 276, 2001
|
|
6CCU
 
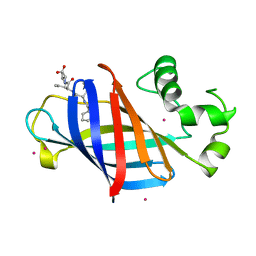 | | Complex between a GID4 fragment and a short peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Short peptide, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CD9
 
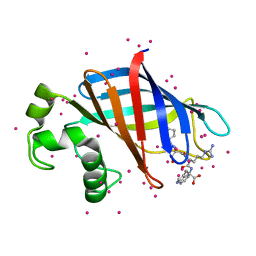 | | GID4 in complex with a peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide PSRW, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CM2
 
 | | SAMHD1 HD domain bound to decitabine triphosphate | | Descriptor: | 6-amino-3-{2-deoxy-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-3,4-dihydro-1,3,5-triazin-2(1H)-one, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Oellerich, T, Schneider, C, Thomas, D, Knecht, K.M, Buzovetsky, O, Kaderali, L, Schliemann, C, Bohnenberger, H, Angenendt, L, Hartmann, W, Wardelmann, E, Rothenburger, T, Mohr, S, Scheich, S, Comoglio, F, Wilke, A, Strobel, P, Serve, H, Michaelis, M, Ferreiros, N, Geisslinger, G, Xiong, Y, Keppler, O.T, Cinatl, J. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Selective inactivation of hypomethylating agents by SAMHD1 provides a rationale for therapeutic stratification in AML.
Nat Commun, 10, 2019
|
|
6CMP
 
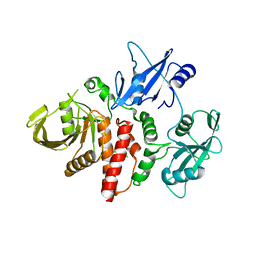 | | Closed structure of inactive SHP2 mutant C459E | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Padua, R.A.P, Sun, Y, Marko, I, Pitsawong, W, Kern, D. | | Deposit date: | 2018-03-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2.
Nat Commun, 9, 2018
|
|
1GDT
 
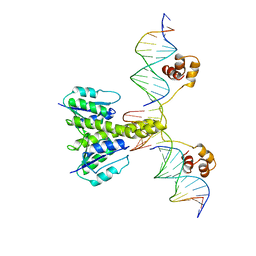 | |
6CCT
 
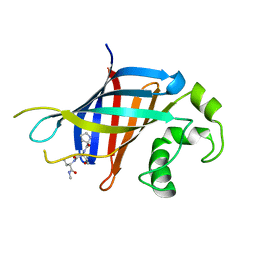 | | Fragment of GID4 in complex with a short peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
1HBU
 
 | |
6CKO
 
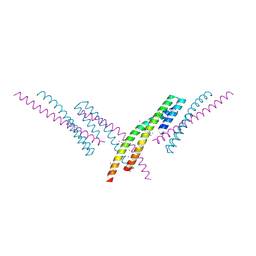 | | Crystal structure of an AF10 fragment | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, Protein AF-10, ... | | Authors: | Zhang, H, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the DOT1L-AF10 complex reveals mechanistic insights into MLL-AF10-associated leukemogenesis.
Genes Dev., 32, 2018
|
|
1GTL
 
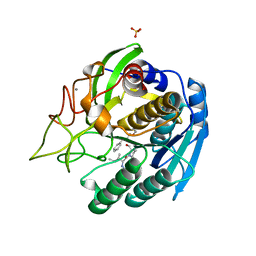 | | The thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Pro-Phe-cho | | Descriptor: | ALDEHYDE INHIBITOR, CALCIUM ION, KUMAMOLYSIN, ... | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-16 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
6CT9
 
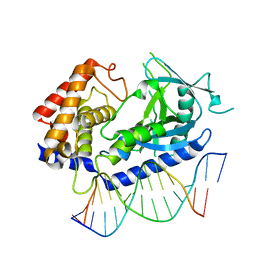 | | Structure of the human cGAS-DNA complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*GP*A)-3'), DNA (5'-D(P*CP*GP*TP*CP*TP*TP*CP*GP*GP*CP*AP*AP*T)-3'), ... | | Authors: | Zhou, W, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-03-22 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of the Human cGAS-DNA Complex Reveals Enhanced Control of Immune Surveillance.
Cell, 174, 2018
|
|
6D0S
 
 | | RabGAP domain of human TBC1D22B | | Descriptor: | SULFATE ION, TBC1 domain family member 22B, UNKNOWN ATOM OR ION | | Authors: | Tong, Y, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RabGAP domain of human TBC1D22B
To be Published
|
|
