3PHD
 
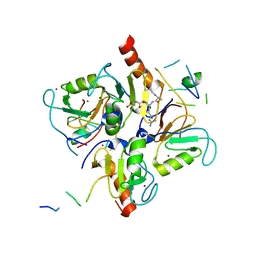 | | Crystal structure of human HDAC6 in complex with ubiquitin | | Descriptor: | Histone deacetylase 6, Polyubiquitin, ZINC ION | | Authors: | Dong, A, Qui, W, Ravichandran, M, Schuetz, A, Loppnau, P, Li, F, Mackenzie, F, Kozieradzki, I, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-03 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Aggregates Are Recruited to Aggresome by Histone Deacetylase 6 via Unanchored Ubiquitin C Termini.
J.Biol.Chem., 287, 2012
|
|
1VHI
 
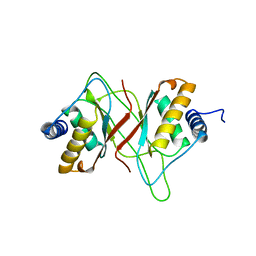 | | EPSTEIN BARR VIRUS NUCLEAR ANTIGEN-1 DNA-BINDING DOMAIN, RESIDUES 470-607 | | Descriptor: | EPSTEIN BARR VIRUS NUCLEAR ANTIGEN-1 | | Authors: | Bochkarev, A, Barwell, J, Pfuetzner, R, Furey, W, Edwards, A, Frappier, L. | | Deposit date: | 1996-10-05 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein EBNA 1.
Cell(Cambridge,Mass.), 83, 1995
|
|
2MRB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD-7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
2MRT
 
 | | CONFORMATION OF CD-7 METALLOTHIONEIN-2 FROM RAT LIVER IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformation of [Cd7]-metallothionein-2 from rat liver in aqueous solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 203, 1988
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
2CUL
 
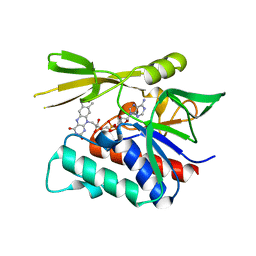 | | Crystal structure of the GidA-related protein from Thermus thermophilus HB8 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glucose-inhibited division protein A-related protein, probable oxidoreductase | | Authors: | Iwasaki, W, Miyatake, H, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-26 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the small form of glucose-inhibited division protein A from Thermus thermophilus HB8
Proteins, 61, 2005
|
|
2R13
 
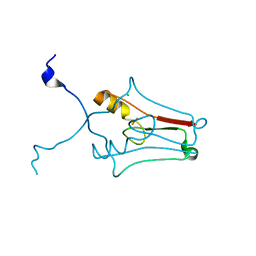 | | Crystal structure of human mitoNEET reveals a novel [2Fe-2S] cluster coordination | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Zinc finger CDGSH domain-containing protein 1 | | Authors: | Hou, X, Liu, R, Ross, S, Smart, E.J, Zhu, H, Gong, W. | | Deposit date: | 2007-08-22 | | Release date: | 2007-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies of human MitoNEET
J.Biol.Chem., 282, 2007
|
|
1DAG
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID AND 5'-ADENOSYL-METHYLENE-TRIPHOSPHATE | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
2QQ8
 
 | | Crystal structure of the putative RabGAP domain of human TBC1 domain family member 14 | | Descriptor: | TBC1 domain family member 14, UNKNOWN ATOM OR ION | | Authors: | Tong, Y, Tempel, W, Dimov, S, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-26 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative RabGAP domain of human TBC1 domain family member 14.
To be Published
|
|
8TF7
 
 | |
2QT5
 
 | | Crystal Structure of GRIP1 PDZ12 in Complex with the Fras1 Peptide | | Descriptor: | (ASN)(ASN)(LEU)(GLN)(ASP)(GLY)(THR)(GLU)(VAL), 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Long, J, Wei, Z, Feng, W, Zhao, Y, Zhang, M. | | Deposit date: | 2007-08-01 | | Release date: | 2008-06-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Supramodular nature of GRIP1 revealed by the structure of its PDZ12 tandem in complex with the carboxyl tail of Fras1.
J.Mol.Biol., 375, 2008
|
|
2R2D
 
 | | Structure of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, PHOSPHATE ION, ZINC ION, ... | | Authors: | Liu, D, Thomas, P.W, Momb, J, Hoang, Q, Petsko, G.A, Ringe, D, Fast, W. | | Deposit date: | 2007-08-24 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and specificity of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens.
Biochemistry, 46, 2007
|
|
2REO
 
 | | Crystal structure of human sulfotransferase 1C3 (Sult1C3) in complex with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Putative sulfotransferase 1C3 | | Authors: | Tempel, W, Pan, P, Dong, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-26 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Crystal structure of human sulfotransferase 1C3 (Sult1C3) in complex with PAP.
To be Published
|
|
2R53
 
 | |
2R9H
 
 | |
8X0J
 
 | | old yellow enzymes-Q102A/Y169F/R215A/R308A | | Descriptor: | NADPH dehydrogenase, PHOSPHATE ION | | Authors: | Lei, W, Wei, S. | | Deposit date: | 2023-11-04 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Unlocking the function promiscuity of old yellow enzyme to catalyze asymmetric Morita-Baylis-Hillman reaction
To Be Published
|
|
1DAH
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7,8-DIAMINO-NONANOIC ACID, 5'-ADENOSYL-METHYLENE-TRIPHOSPHATE, AND MANGANESE | | Descriptor: | 7,8-DIAMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE, MANGANESE (II) ION, ... | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
8XNH
 
 | | Nipah virus fusion glycoprotein in complex with a broadly neutralizing antibody 5C8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5C8-VH, 5C8-VL, ... | | Authors: | Fan, P.F, Ren, Y, Yu, C.M, Chen, W. | | Deposit date: | 2023-12-30 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Nipah virus fusion glycoprotein in complex with a broadly neutralizing antibody 1D6
To Be Published
|
|
1DAI
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
2D8L
 
 | | Crystal Structure of Unsaturated Rhamnogalacturonyl Hydrolase in complex with dGlcA-GalNAc | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, Putative glycosyl hydrolase yteR | | Authors: | Itoh, T, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2005-12-06 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel glycoside hydrolase family 105: the structure of family 105 unsaturated rhamnogalacturonyl hydrolase complexed with a disaccharide in comparison with family 88 enzyme complexed with the disaccharide
J.Mol.Biol., 360, 2006
|
|
2R9M
 
 | | Cathepsin S complexed with Compound 15 | | Descriptor: | Cathepsin S, N-[(1S)-2-[(4-cyano-1-methylpiperidin-4-yl)amino]-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | Authors: | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2007-09-13 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
To be Published
|
|
4GGD
 
 | |
2RGH
 
 | | Structure of Alpha-Glycerophosphate Oxidase from Streptococcus sp.: A Template for the Mitochondrial Alpha-Glycerophosphate Dehydrogenase | | Descriptor: | Alpha-Glycerophosphate Oxidase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Colussi, T, Boles, W, Mallett, T.C, Karplus, P.A, Claiborne, A. | | Deposit date: | 2007-10-01 | | Release date: | 2008-01-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of alpha-glycerophosphate oxidase from Streptococcus sp.: a template for the mitochondrial alpha-glycerophosphate dehydrogenase.
Biochemistry, 47, 2008
|
|
2RG2
 
 | |
121P
 
 | | STRUKTUR UND GUANOSINTRIPHOSPHAT-HYDROLYSEMECHANISMUS DES C-TERMINAL VERKUERZTEN MENSCHLICHEN KREBSPROTEINS P21-H-RAS | | Descriptor: | H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Krengel, U, Scheffzek, K, Scherer, A, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Struktur Und Guanosintriphosphat-Hydrolysemechanismus Des C-Terminal Verkuerzten Menschlichen Krebsproteins P21-H-Ras
Thesis, 1991
|
|
