6RDD
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Primary rotary state 2, monomer-masked refinement | | 分子名称: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ATP synthase associated protein ASA1, ATP synthase subunit alpha, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDN
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1C, monomer-masked refinement | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RE3
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2B, monomer-masked refinement | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6R6T
 
 | | Crystal structure of mouse cis-aconitate decarboxylase | | 分子名称: | Cis-aconitate decarboxylase | | 著者 | Lukat, P, Chen, F, Saile, K, Buessow, K, Pessler, F, Blankenfeldt, W. | | 登録日 | 2019-03-28 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.535 Å) | | 主引用文献 | Crystal structure ofcis-aconitate decarboxylase reveals the impact of naturally occurring human mutations on itaconate synthesis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6REU
 
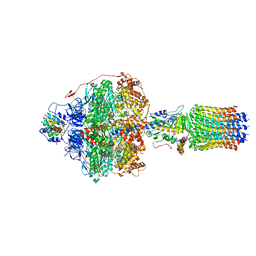 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3C, focussed refinement of F1 head and rotor | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6R36
 
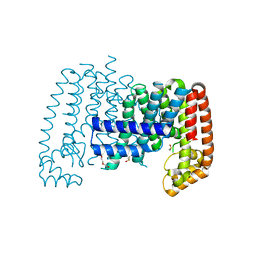 | | T. brucei farnesyl pyrophosphate synthase (FPPS) | | 分子名称: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, Farnesyl pyrophosphate synthase | | 著者 | Muenzker, L, Petrick, J.K, Schleberger, C, Jahnke, W. | | 登録日 | 2019-03-19 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Fragment-Based Discovery of Non-bisphosphonate Binders of Trypanosoma brucei Farnesyl Pyrophosphate Synthase.
Chembiochem, 21, 2020
|
|
6R4R
 
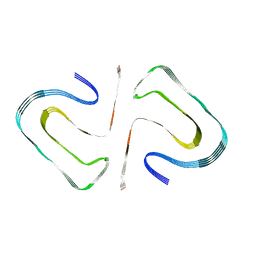 | | Cryo-EM Structure of the PI3-Kinase SH3 Domain Amyloid Fibril | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha | | 著者 | Roeder, C, Vettore, N, Mangels, L.N, Gremer, L, Ravelli, R.B.G, Willbold, D, Hoyer, W, Buell, A.K, Schroder, G.F. | | 登録日 | 2019-03-23 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Atomic structure of PI3-kinase SH3 amyloid fibrils by cryo-electron microscopy.
Nat Commun, 10, 2019
|
|
6R9D
 
 | |
1A6B
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE ZINC FINGER PROTEIN NCP10 OF MOLONEY MURINE LEUKEMIA VIRUS AND A SEQUENCE OF THE PSI-PACKAGING DOMAIN OF HIV-1, 20 STRUCTURES | | 分子名称: | DNA (5'-D(*AP*CP*GP*CP*C)-3'), ZINC FINGER PROTEIN NCP10, ZINC ION | | 著者 | Schueler, W, Dong, C.-Z, Wecker, K, Roques, B.P. | | 登録日 | 1998-02-23 | | 公開日 | 1999-08-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the complex between the zinc finger protein NCp10 of Moloney murine leukemia virus and the single-stranded pentanucleotide d(ACGCC): comparison with HIV-NCp7 complexes.
Biochemistry, 38, 1999
|
|
1OHJ
 
 | | HUMAN DIHYDROFOLATE REDUCTASE, MONOCLINIC (P21) CRYSTAL FORM | | 分子名称: | DIHYDROFOLATE REDUCTASE, N-(4-CARBOXY-4-{4-[(2,4-DIAMINO-PTERIDIN-6-YLMETHYL)-AMINO]-BENZOYLAMINO}-BUTYL)-PHTHALAMIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W. | | 登録日 | 1997-09-17 | | 公開日 | 1998-04-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Comparison of two independent crystal structures of human dihydrofolate reductase ternary complexes reduced with nicotinamide adenine dinucleotide phosphate and the very tight-binding inhibitor PT523.
Biochemistry, 36, 1997
|
|
1OHK
 
 | | HUMAN DIHYDROFOLATE REDUCTASE, ORTHORHOMBIC (P21 21 21) CRYSTAL FORM | | 分子名称: | DIHYDROFOLATE REDUCTASE, N-(4-CARBOXY-4-{4-[(2,4-DIAMINO-PTERIDIN-6-YLMETHYL)-AMINO]-BENZOYLAMINO}-BUTYL)-PHTHALAMIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W. | | 登録日 | 1997-09-17 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Comparison of two independent crystal structures of human dihydrofolate reductase ternary complexes reduced with nicotinamide adenine dinucleotide phosphate and the very tight-binding inhibitor PT523.
Biochemistry, 36, 1997
|
|
6S0S
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, ribostamycin B and 2-oxoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, CHLORIDE ION, Kanamycin B dioxygenase, ... | | 著者 | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6RJR
 
 | | Crystal structure of a Fungal Catalase at 1.9 Angstrom | | 分子名称: | CHLORIDE ION, Catalase, GLYCEROL, ... | | 著者 | Gomez, S, Navas-Yuste, S, Payne, A.M, Rivera, W, Lopez-Estepa, M, Brangbour, C, Fulla, D, Juanhuix, J, Fernandez, F.J, Vega, M.C. | | 登録日 | 2019-04-29 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.895 Å) | | 主引用文献 | Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes.
Free Radic. Biol. Med., 141, 2019
|
|
2BTC
 
 | | BOVINE TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA PEPO TRYPSIN INHIBITOR II) | | 分子名称: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | 著者 | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | 登録日 | 1998-12-11 | | 公開日 | 2000-01-19 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6XQB
 
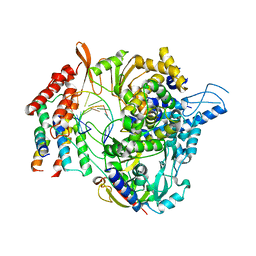 | | SARS-CoV-2 RdRp/RNA complex | | 分子名称: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Liu, B, Shi, W, Yang, Y. | | 登録日 | 2020-07-09 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of SARS-CoV-2 RdRp/RNA complex at 3.4 Angstroms resolution
To Be Published
|
|
6S0W
 
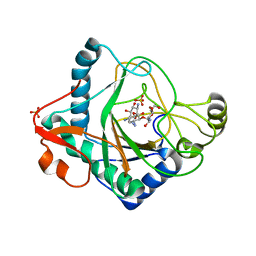 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel and kanamycin B sulfate | | 分子名称: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, DI(HYDROXYETHYL)ETHER, Kanamycin B dioxygenase, ... | | 著者 | Mrugala, B, Niedzialkowska, E, Minor, W, Borowski, T. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6XUY
 
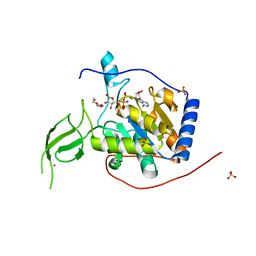 | |
6RJN
 
 | | Crystal structure of a Fungal Catalase at 2.3 Angstroms | | 分子名称: | CHLORIDE ION, Catalase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Gomez, S, Navas-Yuste, S, Payne, A.M, Rivera, W, Lopez-Estepa, M, Brangbour, C, Fulla, D, Juanhuix, J, Fernandez, F.J, Vega, M.C. | | 登録日 | 2019-04-28 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.295 Å) | | 主引用文献 | Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes.
Free Radic. Biol. Med., 141, 2019
|
|
6ROJ
 
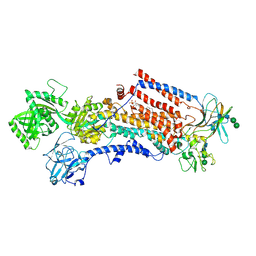 | | Cryo-EM structure of the activated Drs2p-Cdc50p | | 分子名称: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell division control protein 50, ... | | 著者 | Timcenko, M, Lyons, J.A, Januliene, D, Ulstrup, J.J, Dieudonne, T, Montigny, C, Ash, M.R, Karlsen, J.L, Boesen, T, Kuhlbrandt, W, Lenoir, G, Moeller, A, Nissen, P. | | 登録日 | 2019-05-13 | | 公開日 | 2019-07-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure and autoregulation of a P4-ATPase lipid flippase.
Nature, 571, 2019
|
|
6XB9
 
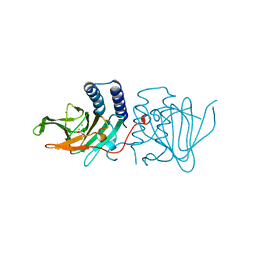 | | Crystal structure of Azotobacter vinelandii 3-mercaptopropionic acid dioxygenase in complex with 3-hydroxypropionic acid | | 分子名称: | 3-HYDROXY-PROPANOIC ACID, CHLORIDE ION, Cysteine dioxygenase type I protein, ... | | 著者 | Kiser, P.D, Khadka, N, Shi, W, Pierce, B.S. | | 登録日 | 2020-06-05 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure of 3-mercaptopropionic acid dioxygenase with a substrate analog reveals bidentate substrate binding at the iron center.
J.Biol.Chem., 296, 2021
|
|
1B64
 
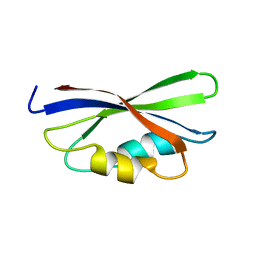 | | SOLUTION STRUCTURE OF THE GUANINE NUCLEOTIDE EXCHANGE FACTOR DOMAIN FROM HUMAN ELONGATION FACTOR-ONE BETA, NMR, 20 STRUCTURES | | 分子名称: | ELONGATION FACTOR 1-BETA | | 著者 | Perez, J.M.J, Siegal, G, Kriek, J, Hard, K, Dijk, J, Canters, G.W, Moller, W. | | 登録日 | 1999-01-20 | | 公開日 | 1999-05-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the guanine nucleotide exchange domain of human elongation factor 1beta reveals a striking resemblance to that of EF-Ts from Escherichia coli.
Structure Fold.Des., 7, 1999
|
|
1B8O
 
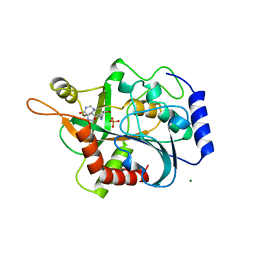 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | 分子名称: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Fedorov, A.A, Kicska, G.A, Fedorov, E.V, Shi, W, Tyler, P.C, Furneaux, R.H, Schramm, V.L, Almo, S.C. | | 登録日 | 1999-02-02 | | 公開日 | 1999-02-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis.
Biochemistry, 40, 2001
|
|
6ROI
 
 | | Cryo-EM structure of the partially activated Drs2p-Cdc50p | | 分子名称: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Timcenko, M, Lyons, J.A, Januliene, D, Ulstrup, J.J, Dieudonne, T, Montigny, C, Ash, M.R, Karlsen, J.L, Boesen, T, Kuhlbrandt, W, Lenoir, G, Moeller, A, Nissen, P. | | 登録日 | 2019-05-13 | | 公開日 | 2019-07-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure and autoregulation of a P4-ATPase lipid flippase.
Nature, 571, 2019
|
|
6XDF
 
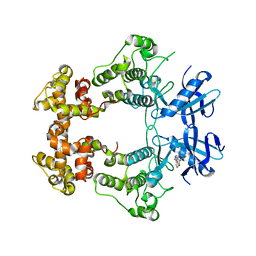 | | Crystal structure of IRE1a in complex with G-4100 | | 分子名称: | 4-amino-N-(6-chloro-2-fluoro-3-{[(pyrrolidin-1-yl)sulfonyl]amino}phenyl)quinazoline-8-carboxamide, SODIUM ION, Serine/threonine-protein kinase/endoribonuclease IRE1 | | 著者 | Steinbacher, S, Wang, W. | | 登録日 | 2020-06-10 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Identification of BRaf-Sparing Amino-Thienopyrimidines with Potent IRE1 alpha Inhibitory Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
6XDD
 
 | | Crystal structure of IRE1 in complex with G-3053 | | 分子名称: | 4-[(trans-4-aminocyclohexyl)amino]-N-(6-chloro-3-{[(2,5-difluorophenyl)sulfonyl]amino}-2-fluorophenyl)thieno[3,2-d]pyrimidine-7-carboxamide, Serine/threonine-protein kinase/endoribonuclease IRE1 | | 著者 | Ackerly-Wallweber, H, Wang, W. | | 登録日 | 2020-06-10 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Identification of BRaf-Sparing Amino-Thienopyrimidines with Potent IRE1 alpha Inhibitory Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
