6CXD
 
 | | Crystal structure of peptidase B from Yersinia pestis CO92 at 2.75 A resolution | | Descriptor: | Peptidase B, SULFATE ION | | Authors: | Woinska, M, Lipowska, J, Shabalin, I.G, Cymborowski, M, Grimshaw, S, Winsor, J, Shuvalova, L, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-02 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
1G2Q
 
 | |
6CCX
 
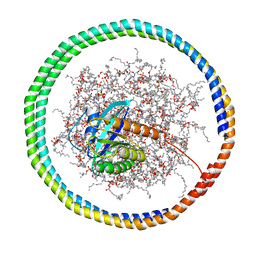 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
1GHV
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-OXO-1,2-DIHYDRO-PYRIDIN-3-YL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, SODIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI9
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-5-METHOXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI2
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-PHENYL)-3H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GTJ
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Ala-Phe-cho | | Descriptor: | ALDEHYDE INHIBITOR, CALCIUM ION, KUMAMOLYSIN, ... | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-15 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GTP
 
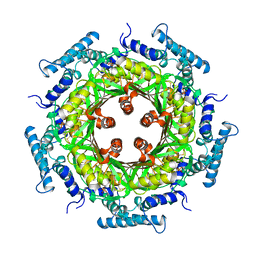 | | GTP CYCLOHYDROLASE I | | Descriptor: | GTP CYCLOHYDROLASE I, SULFATE ION | | Authors: | Nar, H, Huber, R, Meining, W, Bacher, A. | | Deposit date: | 1995-09-16 | | Release date: | 1996-11-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Atomic structure of GTP cyclohydrolase I.
Structure, 3, 1995
|
|
1H4U
 
 | | Domain G2 of mouse nidogen-1 | | Descriptor: | NIDOGEN-1 | | Authors: | Hopf, M, Gohring, W, Ries, A, Timpl, R, Hohenester, E. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure and Mutational Analysis of a Perlecan-Binding Fragment of Nidogen-1
Nat.Struct.Biol., 8, 2001
|
|
1H5X
 
 | |
7X4L
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase mutant Y303F-PLP complex | | Descriptor: | Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Liu, S, Guoming, D, Yulu, W, Boting, W, Xin, F. | | Deposit date: | 2022-03-02 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
1FYD
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH ONE MOLECULE AMP, ONE PYROPHOSPHATE ION AND ONE MG2+ ION | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) SYNTHETASE, ... | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Brouillette, W, Muccio, D, Jedrzejas, M, Brouillette, C, DeLucas, L. | | Deposit date: | 2000-09-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FU0
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE PHOSPHO-SERINE 46 HPR FROM ENTEROCOCCUS FAECALIS | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR | | Authors: | Audette, G.F, Engelmann, R, Hengstenberg, W, Deutscher, J, Hayakawa, K, Quail, J.W, Delbaere, L.T.J. | | Deposit date: | 2000-09-13 | | Release date: | 2000-11-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A resolution structure of phospho-serine 46 HPr from Enterococcus faecalis.
J.Mol.Biol., 303, 2000
|
|
1G27
 
 | | CRYSTAL STRUCTURE OF E.COLI POLYPEPTIDE DEFORMYLASE COMPLEXED WITH THE INHIBITOR BB-3497 | | Descriptor: | 2-[(FORMYL-HYDROXY-AMINO)-METHYL]-HEXANOIC ACID (1-DIMETHYLCARBAMOYL-2,2-DIMETHYL-PROPYL)-AMIDE, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-17 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
1G2P
 
 | | CRYSTAL STRUCTURE OF ADENINE PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | ADENINE PHOSPHORIBOSYLTRANSFERASE 1, SULFATE ION | | Authors: | Shi, W, Tanaka, K.S.E, Almo, S.C, Schramm, V.L. | | Deposit date: | 2000-10-20 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of adenine phosphoribosyltransferase from Saccharomyces cerevisiae.
Biochemistry, 40, 2001
|
|
1G4K
 
 | | X-ray Structure of a Novel Matrix Metalloproteinase Inhibitor Complexed to Stromelysin | | Descriptor: | 5-METHYL-5-(4-PHENOXY-PHENYL)-PYRIMIDINE-2,4,6-TRIONE, CALCIUM ION, GLYCEROL, ... | | Authors: | Dunten, P, Kammlott, U, Crowther, R, Levin, W, Foley, L.H, Wang, P, Palermo, R. | | Deposit date: | 2000-10-27 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of a novel matrix metalloproteinase inhibitor complexed to stromelysin.
Protein Sci., 10, 2001
|
|
1G8Y
 
 | | CRYSTAL STRUCTURE OF THE HEXAMERIC REPLICATIVE HELICASE REPA OF PLASMID RSF1010 | | Descriptor: | REGULATORY PROTEIN REPA | | Authors: | Niedenzu, T, Roeleke, D, Bains, G, Scherzinger, E, Saenger, W. | | Deposit date: | 2000-11-21 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010.
J.Mol.Biol., 306, 2001
|
|
1FW6
 
 | | CRYSTAL STRUCTURE OF A TAQ MUTS-DNA-ADP TERNARY COMPLEX | | Descriptor: | 5'-D(*GP*CP*GP*AP*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*TP*CP*GP*TP*C)-3', 5'-D(*GP*GP*AP*CP*GP*AP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*GP*TP*CP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Junop, M.S, Obmolova, G, Rausch, K, Hsieh, P, Yang, W. | | Deposit date: | 2000-09-21 | | Release date: | 2001-02-19 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair.
Mol.Cell, 7, 2001
|
|
1HBM
 
 | | METHYL-COENZYME M REDUCTASE ENZYME PRODUCT COMPLEX | | Descriptor: | CHLORIDE ION, FACTOR 430, GLYCEROL, ... | | Authors: | Ermler, U, Grabarse, W. | | Deposit date: | 2001-04-20 | | Release date: | 2001-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | On the Mechanism of Biological Methane Formation: Structural Evidence for Conformational Changes in Methyl-Coenzyme M Reductase Upon Substrate Binding
J.Mol.Biol., 309, 2001
|
|
1GTG
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolysin (kscp) | | Descriptor: | CALCIUM ION, KUMAMOLYSIN | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-15 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GYX
 
 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BENZOIC ACID, HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-30 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
1H30
 
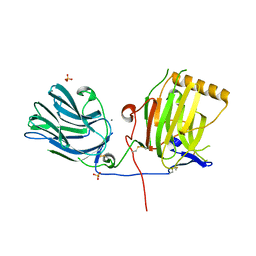 | | C-terminal LG domain pair of human Gas6 | | Descriptor: | CALCIUM ION, GROWTH-ARREST-SPECIFIC PROTEIN, SULFATE ION | | Authors: | Sasaki, T, Knyazev, P.G, Cheburkin, Y, Gohring, W, Tisi, D, Ullrich, A, Timpl, R, Hohenester, E. | | Deposit date: | 2002-08-21 | | Release date: | 2003-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Carboxy-Terminal Fragment of Growth-Arrest-Specific Protein Gas6: Receptor Tyrosine Kinase Activation by Laminin G-Like Domains
J.Biol.Chem., 277, 2002
|
|
1GV3
 
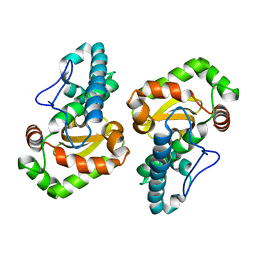 | | The 2.0 Angstrom resolution structure of the catalytic portion of a cyanobacterial membrane-bound manganese superoxide dismutase | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Atzenhofer, W, Regelsberger, G, Jacob, U, Huber, R, Peschek, G.A, Obinger, C. | | Deposit date: | 2002-02-05 | | Release date: | 2002-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0A Resolution Structure of the Catalytic Portion of a Cyanobacterial Membrane-Bound Manganese Superoxide Dismutase
J.Mol.Biol., 321, 2002
|
|
1FXY
 
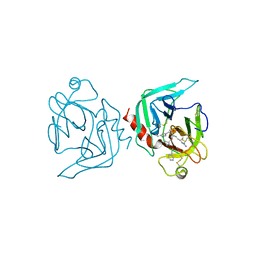 | | COAGULATION FACTOR XA-TRYPSIN CHIMERA INHIBITED WITH D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | COAGULATION FACTOR XA-TRYPSIN CHIMERA, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide | | Authors: | Hopfner, K.P, Kopetzki, E, Kresse, G.-B, Huber, R, Bode, W, Engh, R.A. | | Deposit date: | 1998-04-22 | | Release date: | 1998-06-17 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | New enzyme lineages by subdomain shuffling.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
