6NLZ
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment degradation product B9D | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, [(1R,2S)-2-(2-hydroxybenzene-1-carbonyl)cyclopentyl]acetic acid, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-10 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment degradation product B9D
To Be Published
|
|
1XUA
 
 | | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, Phenazine biosynthesis protein phzF | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4IRR
 
 | | Crystal Structure of C.elegans Thymidylate Synthase in Complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of C.elegans Thymidylate Synthase in Complex with dUMP
To be Published
|
|
6NN0
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with 2'-deoxycytidine and fragment degradation product B9D | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with 2'-deoxycytidine and B9D
To Be Published
|
|
5TA0
 
 | | Crystal structure of BuGH86E322Q in complex with neoagarooctaose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Boraston, A.B, Abbott, W.D. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
1XI8
 
 | | Molybdenum cofactor biosynthesis protein from Pyrococcus furiosus Pfu-1657500-001 | | Descriptor: | Molybdenum cofactor biosynthesis protein | | Authors: | Zhou, W, Zhao, M, Chang, J.C, Liu, Z.-J, Chen, L, Horanyi, P, Xu, H, Yang, H, Habel, J.E, Lee, D, Chang, S.-H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Molybdenum cofactor biosynthesis protein from Pyrococcus furiosus Pfu-1657500-001
To be published
|
|
2FKZ
 
 | | Reduced (All Ferrous) form of the Azotobacter vinelandii bacterioferritin | | Descriptor: | Bacterioferritin, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Swartz, L, Kunchinskas, M, Li, H, Poulos, T.L, Lanzilotta, W.N. | | Deposit date: | 2006-01-05 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-Dependent Structural Changes in the Azotobacter vinelandii Bacterioferritin: New Insights into the Ferroxidase and Iron Transport Mechanism(,).
Biochemistry, 45, 2006
|
|
4XPL
 
 | | The crystal structure of Campylobacter jejuni N-acetyltransferase PseH in complex with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, N-Acetyltransferase, PseH | | Authors: | Song, W.S, Nam, M.S, Namgung, B, Yoon, S.I. | | Deposit date: | 2015-01-17 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of PseH, the Campylobacter jejuni N-acetyltransferase involved in bacterial O-linked glycosylation.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
2ARU
 
 | | Crystal structure of lipoate-protein ligase A bound with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipoate-protein ligase A, MAGNESIUM ION | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
4XZ8
 
 | | The crystal structure of Erve virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
5LY6
 
 | | CryoEM structure of the membrane pore complex of Pneumolysin at 4.5A | | Descriptor: | Pneumolysin | | Authors: | van Pee, K, Neuhaus, A, D'Imprima, E, Mills, D.J, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2016-09-24 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin.
Elife, 6, 2017
|
|
4Y02
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis (Ground) | | Descriptor: | GLYCEROL, POTASSIUM ION, Peptidase S46 | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
4XVR
 
 | | H-Ras Y137F | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2015-01-27 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Tyrosine phosphorylation of RAS by ABL allosterically enhances effector binding.
Faseb J., 29, 2015
|
|
4J4V
 
 | | Pentameric SFTSVN with suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Nucleocapsid protein | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
2F2G
 
 | | X-Ray Structure of Gene Product From Arabidopsis Thaliana AT3G16990 | | Descriptor: | 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, SEED MATURATION PROTEIN PM36 HOMOLOG, SULFATE ION | | Authors: | Wesenberg, G.W, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-11-16 | | Release date: | 2005-12-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of gene locus At3g16990 from Arabidopsis thaliana
Proteins, 57, 2004
|
|
5THS
 
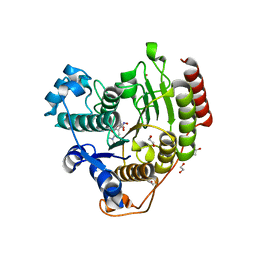 | | Crystal Structure of G302A HDAC8 in complex with M344 | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2016-09-30 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
6O26
 
 | | Crystal structure of 3246 Fab in complex with circumsporozoite protein NANA | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3246 Fab heavy chain, ... | | Authors: | Scally, S.W, Bosch, A, Castro, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-03-04 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
1JGE
 
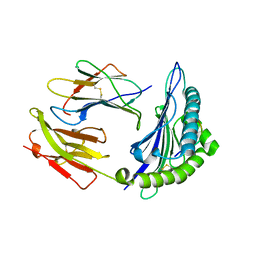 | | HLA-B*2705 bound to nona-peptide m9 | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-27 B*2705 ALPHA CHAIN, ... | | Authors: | Hulsmeyer, M, Hillig, R.C, Volz, A, Ruhl, M, Schroder, W, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2001-06-25 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | HLA-B27 subtypes differentially associated with disease exhibit subtle structural
alterations
J.Biol.Chem., 277, 2002
|
|
3ZHH
 
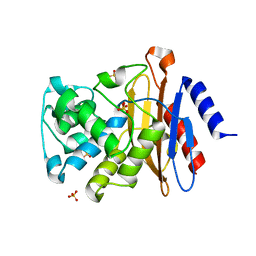 | | X-ray structure of the full-length beta-lactamase from M.tuberculosis | | Descriptor: | BETA-LACTAMASE, SULFATE ION | | Authors: | Feiler, C, Fisher, A.C, Marrichi, M.J, Wright, L, Schmidpeter, P.A.M, Blankenfeldt, W, Pavelka, M, DeLisa, M.P. | | Deposit date: | 2012-12-21 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Directed Evolution of Mycobacterium Tuberculosis Beta-Lactamase Reveals Gatekeeper Residue that Regulates Antibiotic Resistance and Catalytic Efficiency.
Plos One, 8, 2013
|
|
1JDY
 
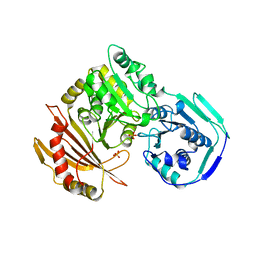 | |
4XSU
 
 | | Crystal structure of Anabaena Alr3699/HepE in complex with UDP and glucose | | Descriptor: | Alr3699 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Wang, X.P, Dai, Y.N, Jiang, Y.L, Cheng, W, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and enzymatic analyses of a glucosyltransferase Alr3699/HepE involved in Anabaena heterocyst envelop polysaccharide biosynthesis
Glycobiology, 26, 2016
|
|
2AV3
 
 | | F97L- no ligand | | Descriptor: | Globin I, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Bonham, M.A, Gibson, Q.H, Nichols, J.C, Royer Jr, W.E. | | Deposit date: | 2005-08-29 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Residue F4 plays a key role in modulating oxygen affinity and cooperativity in Scapharca dimeric hemoglobin
Biochemistry, 44, 2005
|
|
3MJZ
 
 | | The crystal structure of native FG41 MSAD | | Descriptor: | FG41 Malonate Semialdehyde Decarboxylase | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson, W.H.Jr, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-13 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
4WX8
 
 | | Crystal structure of binary complex Gon7-Pcc1 | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit GON7, EKC/KEOPS complex subunit PCC1 | | Authors: | Zhang, W, Van Tilbeurgh, H. | | Deposit date: | 2014-11-13 | | Release date: | 2015-03-18 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
1JCA
 
 | | Non-standard Design of Unstable Insulin Analogues with Enhanced Activity | | Descriptor: | ZINC ION, insulin a, insulin b | | Authors: | Weiss, M.A, Wan, Z, Zhao, M, Chu, Y.-C, Nakagawa, S.H, Burke, G.T, Jia, W, Hellmich, R, Katsoyannis, P.G. | | Deposit date: | 2001-06-08 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor.
J.Mol.Biol., 315, 2002
|
|
