4UIE
 
 | | Crystal structure of the S-layer protein SbsC, domains 7, 8 and 9 | | Descriptor: | CALCIUM ION, OSMIUM ION, SURFACE LAYER PROTEIN | | Authors: | Pavkov-Keller, T, Dordic, A, Egelseer, E.M, Sleytr, U.B, Keller, W. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the S-Layer Protein Sbsc
To be Published
|
|
4PNA
 
 | | A de novo designed heptameric coiled coil CC-Hept | | Descriptor: | CC-Hept, GLYCEROL | | Authors: | Burton, A.J, Wood, C.W, Thomson, A.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-22 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computational design of water-soluble alpha-helical barrels.
Science, 346, 2014
|
|
3D6R
 
 | |
3MFM
 
 | | Crystal Structures and Mutational Analyses of Acyl-CoA Carboxylase Subunit of Streptomyces coelicolor | | Descriptor: | Propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Arabolaza, A, Shillito, E.M, Lin, T.-W, Mitchell, D.L, Melgar, M.M. | | Deposit date: | 2010-04-02 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor.
Biochemistry, 49, 2010
|
|
3CTN
 
 | | STRUCTURE OF CALCIUM-SATURATED CARDIAC TROPONIN C, NMR, 30 STRUCTURES | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Sia, S.K, Li, M.X, Spyracopoulos, L, Gagne, S.M, Liu, W, Putkey, J.A, Sykes, B.D. | | Deposit date: | 1997-05-08 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain.
J.Biol.Chem., 272, 1997
|
|
3MKT
 
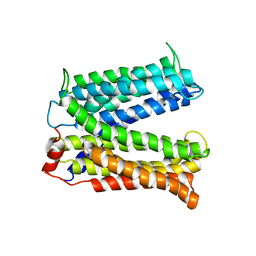 | | Structure of a Cation-bound Multidrug and Toxin Compound Extrusion (MATE) transporter | | Descriptor: | Multi antimicrobial extrusion protein (Na(+)/drug antiporter) MATE-like MDR efflux pump | | Authors: | He, X, Szewczyk, P, Karyakin, A, Evin, M, Hong, W.-X, Zhang, Q, Chang, G. | | Deposit date: | 2010-04-15 | | Release date: | 2010-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structure of a cation-bound multidrug and toxic compound extrusion transporter.
Nature, 467, 2010
|
|
4PFB
 
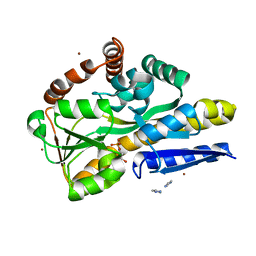 | | Crystal structure of a TRAP periplasmic solute binding protein from Fusobacterium nucleatun (FN1258, TARGET EFI-510120) with bound SN-glycerol-3-phosphate | | Descriptor: | C4-dicarboxylate-binding protein, IMIDAZOLE, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4V4U
 
 | | The quasi-atomic model of Human Adenovirus type 5 capsid | | Descriptor: | HEXON PROTEIN, N-TERMINAL PEPTIDE OF FIBER PROTEIN, PENTON PROTEIN | | Authors: | Fabry, C.M.S, Rosa-Calatrava, M, Conway, J.F, Zubieta, C, Cusack, S, Ruigrok, R.W.H, Schoehn, G. | | Deposit date: | 2005-03-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | A Quasi-Atomic Model of Human Adenovirus Type 5 Capsid.
Embo J., 24, 2005
|
|
2GDC
 
 | | Structure of Vinculin VD1 / IpaA560-633 complex | | Descriptor: | Invasin ipaA, Vinculin | | Authors: | Hamiaux, C, van Eerde, A, Parsot, C, Broos, J, Dijkstra, B.W. | | Deposit date: | 2006-03-15 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural mimicry for vinculin activation by IpaA, a virulence factor of Shigella flexneri.
Embo Rep., 7, 2006
|
|
3MR6
 
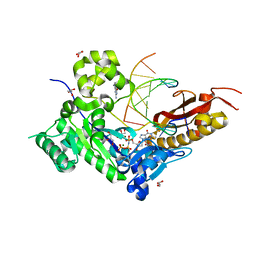 | | Human DNA polymerase eta - DNA ternary complex with a CPD 2bp upstream of the active site (TT4) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*C*AP*TP*CP*AP*(TTD)P*AP*CP*GP*AP*GP*C)-3'), DNA (5'-D(*TP*CP*TP*CP*GP*TP*AP*AP*T)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
3M38
 
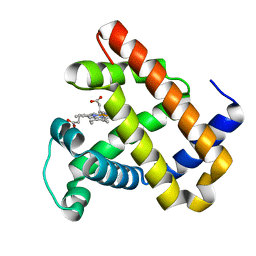 | | The roles of Glutamates and Metal ions in a rationally designed nitric oxide reductase based on myoglobin: I107E FeBMb (No metal ion binding to FeB site) | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lin, Y.-W, Yeung, N, Gao, Y.-G, Miner, K.D, Tian, S, Robinson, H, Lu, Y. | | Deposit date: | 2010-03-08 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2G4D
 
 | | Crystal structure of human SENP1 mutant (C603S) in complex with SUMO-1 | | Descriptor: | SENP1 protein, Small ubiquitin-related modifier 1 | | Authors: | Xu, Z, Chau, S.F, Lam, K.H, Au, S.W.N. | | Deposit date: | 2006-02-22 | | Release date: | 2006-10-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the SENP1 mutant C603S-SUMO complex reveals the hydrolytic mechanism of SUMO-specific protease
Biochem.J., 398, 2006
|
|
2G56
 
 | |
4PGN
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM DESULFOVIBRIO ALASKENSIS G20 (Dde_0634, TARGET EFI-510120) WITH BOUND INDOLE PYRUVATE | | Descriptor: | 1,2-ETHANEDIOL, 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, Extracellular solute-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3M8J
 
 | |
3DD5
 
 | | Glomerella cingulata E600-cutinase complex | | Descriptor: | Cutinase, DIETHYL PHOSPHONATE | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-05 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
4PND
 
 | | A de novo designed pentameric coiled coil CC-Pent_Variant | | Descriptor: | CC-Pent_Variant | | Authors: | Wood, C.W, Burton, A.J, Thomson, A.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-22 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Computational design of water-soluble alpha-helical barrels.
Science, 346, 2014
|
|
3MF7
 
 | | Crystal Structure of (R)-oxirane-2-carboxylate inhibited cis-CaaD | | Descriptor: | Cis-3-chloroacrylic acid dehalogenase | | Authors: | Guo, Y, Serrano, H, Ernst, S.R, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of native and inactivated cis-3-chloroacrylic acid dehalogenase: Implications for the catalytic and inactivation mechanisms.
Bioorg.Chem., 39, 2011
|
|
4POZ
 
 | | Fab fragment of Der p 1 specific antibody 10B9 | | Descriptor: | heavy chain of Fab fragment of 10B9 antibody, light chain of Fab fragment of 10B9 antibody | | Authors: | Osinski, T, Majorek, K.A, Pomes, A, Offermann, L.R, Osinski, S, Glesner, J, Vailes, L.D, Chapman, M.D, Minor, W, Chruszcz, M. | | Deposit date: | 2014-02-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Analysis of Der p 1-Antibody Complexes and Comparison with Complexes of Proteins or Peptides with Monoclonal Antibodies.
J. Immunol., 195, 2015
|
|
3DAE
 
 | |
3MOM
 
 | | Structure of holo HasAp H32A mutant complexed with imidazole from Pseudomonas aeruginosa to 2.25A Resolution | | Descriptor: | Heme acquisition protein HasAp, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Jepkorir, G, Rodriguez, J.C, Rui, H, Im, W, Alontaga, A.Y, Yukl, E, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2010-04-22 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural, NMR Spectroscopic, and Computational Investigation of Hemin Loading in the Hemophore HasAp from Pseudomonas aeruginosa.
J.Am.Chem.Soc., 132, 2010
|
|
3MR2
 
 | | Human DNA polymerase eta in complex with normal DNA and incoming nucleotide (Nrm) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*T*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
2GH9
 
 | | Thermus thermophilus maltotriose binding protein bound with maltotriose | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose/maltodextrin-binding protein | | Authors: | Cuneo, M.J, Changela, A, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2006-03-27 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural adaptations that modulate monosaccharide, disaccharide, and trisaccharide specificities in periplasmic maltose-binding proteins.
J.Mol.Biol., 389, 2009
|
|
3DCN
 
 | | Glomerella cingulata apo cutinase | | Descriptor: | Cutinase | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-04 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
4PGP
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM DESULFOVIBRIO ALASKENSIS G20 (Dde_0634, TARGET EFI-510120) WITH BOUND 3-INDOLE ACETIC ACID | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOL-3-YLACETIC ACID, Extracellular solute-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-05-02 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
