3USY
 
 | |
4YEF
 
 | | beta1 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclododextrin | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-1, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL, ... | | Authors: | Mobbs, J, Gorman, M.A, Parker, M.W, Gooley, P.R, Griffin, M. | | Deposit date: | 2015-02-24 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Determinants of oligosaccharide specificity of the carbohydrate-binding modules of AMP-activated protein kinase.
Biochem.J., 468, 2015
|
|
2OMZ
 
 | |
8P2A
 
 | | Crystal structure of SaFMN3 domain 2 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-binding domain-containing protein | | Authors: | Rozeboom, H.J, Fraaije, M.W. | | Deposit date: | 2023-05-15 | | Release date: | 2023-07-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of two bacterial multi-flavinylated proteins harboring multiple covalent flavin cofactors.
Bba Adv, 4, 2023
|
|
3AZZ
 
 | | Crystal structure of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with gluconolactone | | Descriptor: | CALCIUM ION, D-glucono-1,5-lactone, Laminarinase, ... | | Authors: | Jeng, W.Y, Wang, N.C, Wang, A.H.J. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with inhibitors: essential residues for beta-1,3 and beta-1,4 glucan selection.
J.Biol.Chem., 286, 2011
|
|
6HGB
 
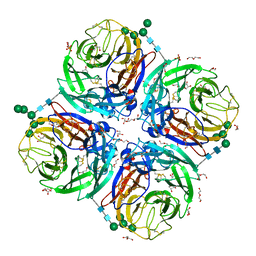 | | Influenza A virus N6 neuraminidase native structure (Duck/England/56). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-23 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
5NEY
 
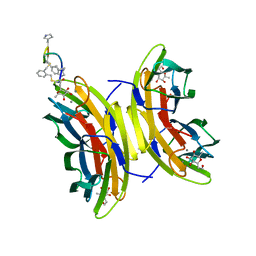 | | Discovery, crystal structures and atomic force microscopy study of thioether ligated D,L-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, CYS-TRD-TRP-LYD-LYS-LYD-LYS-LYD-TRP-TRD-CYS-ALA, ... | | Authors: | Reymond, J.-L, Darbre, T, Stocker, A, Hong, W, van Delden, C, Koehler, T, Luscher, A, Visini, R, Fu, Y, Di Bonaventura, I, He, R. | | Deposit date: | 2017-03-13 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, crystal structure and atomic force microscopy study of thioether ligated d,l-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa.
Chem Sci, 8, 2017
|
|
1W3E
 
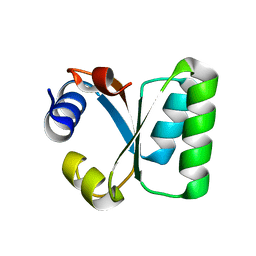 | | Ribosomal L30e of Thermococcus celer, P59A mutant | | Descriptor: | 50S RIBOSOMAL PROTEIN L30E | | Authors: | Ma, H.W, Lee, C.F, Allen, M.D, Bycroft, M, Wong, K.B. | | Deposit date: | 2004-07-15 | | Release date: | 2006-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Role of Proline Residues in Thermostability of T. Celer L30E Protein
To be Published
|
|
6PZR
 
 | |
4EMH
 
 | | Crystal structure of SpLsm4 | | Descriptor: | Probable U6 snRNA-associated Sm-like protein LSm4 | | Authors: | Jiang, S.M, Wu, D.H, Song, H.W. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Lsm3, Lsm4 and Lsm5/6/7 from Schizosaccharomyces pombe.
Plos One, 7, 2012
|
|
5YOF
 
 | |
2JCG
 
 | | Apo form of the catabolite control protein A (ccpA) from bacillus megaterium, with the DNA binding domain | | Descriptor: | CALCIUM ION, GLUCOSE-RESISTANCE AMYLASE REGULATOR | | Authors: | Singh, R.K, Panjikar, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2006-12-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Apo Form of the Catabolite Control Protein a (Ccpa) from Bacillus Megaterium with a DNA-Binding Domain.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2YKU
 
 | | Structural Determinants of the Beta-Selectivity of a Bacterial Aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, BETA-TRANSAMINASE, GLYCEROL, ... | | Authors: | Wybenga, G.G, Crismaru, C.G, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2011-05-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Determinants of the Beta-Selectivity of a Bacterial Aminotransferase.
J.Biol.Chem., 287, 2012
|
|
8J7C
 
 | | Crystal structure of triosephosphate isomerase from Leishmania orientalis at 1.88A with an arsenic ion bound at Cys57 | | Descriptor: | ARSENIC, Triosephosphate isomerase | | Authors: | Kuaprasert, B, Attarataya, J, Riangrungroj, P, Pornthanakasem, W, Suginta, W, Mungthin, M, Leelayoova, S, Choowongkomon, K, Leartsakulpanich, U. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Leishmania orientalis triosephosphate isomerase crystal structure at 1.45 angstroms resolution and its potential specific inhibitors
To be published
|
|
5Y15
 
 | | Crystal structure of human DUSP28 | | Descriptor: | Dual specificity phosphatase 28, PHOSPHATE ION | | Authors: | Ku, B, Hong, W, Kim, S.J, Ryu, S.E. | | Deposit date: | 2017-07-19 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical analysis of atypically low dephosphorylating activity of human dual-specificity phosphatase 28
PLoS ONE, 12, 2017
|
|
8R6S
 
 | | Plastid-encoded RNA polymerase (Integrated model) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-11-22 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
4E45
 
 | | Crystal structure of the hMHF1/hMHF2 Histone-Fold Tetramer in Complex with Fanconi Anemia Associated Helicase hFANCM | | Descriptor: | Centromere protein S, Centromere protein X, Fanconi anemia group M protein, ... | | Authors: | Fox III, D, Zhao, Y, Yang, W, Weidong, W. | | Deposit date: | 2012-03-12 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Reveal that FANCM remodels the MHF Tetramer in favor of binding Branched DNA
To be Published
|
|
8RDJ
 
 | | Plastid-encoded RNA polymerase transcription elongation complex (Integrated model) | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
1YN7
 
 | | Crystal structure of a mouse MHC class I protein, H2-Db, in complex with a mutated peptide (R7A) of the influenza A acid polymerase | | Descriptor: | 10-mer peptide from RNA-directed RNA polymerase subunit P2, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Turner, S.J, Kedzierska, K, Komodromou, H, La Gruta, N.L, Dunstone, M.A, Webb, A.I, Webby, R, Walden, H, Xie, W, McCluskey, J, Purcell, A.W, Rossjohn, J, Doherty, P.C. | | Deposit date: | 2005-01-24 | | Release date: | 2005-06-28 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lack of prominent peptide-major histocompatibility complex features limits repertoire diversity in virus-specific CD8+ T cell populations
Nat.Immunol., 6, 2005
|
|
2JD6
 
 | |
2OMT
 
 | | Crystal structure of InlA G194S+S/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin; E-Cad/CTF1, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamically reengineering the listerial invasion complex InlA/E-cadherin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6VCC
 
 | | Cryo-EM structure of the Dvl2 DIX filament | | Descriptor: | Segment polarity protein dishevelled homolog DVL-2 | | Authors: | Enos, M, Kan, W, Muennich, S, Chen, D.H, Skiniotis, G, Weis, W.I. | | Deposit date: | 2019-12-20 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Limited Dishevelled/Axin oligomerization determines efficiency of Wnt/ beta-catenin signal transduction.
Elife, 9, 2020
|
|
8RAS
 
 | | Plastid-encoded RNA polymerase transcription elongation complex | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
6JBS
 
 | | Bifunctional xylosidase/glucosidase LXYL | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, W.M, Yang, L.Y. | | Deposit date: | 2019-01-26 | | Release date: | 2020-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of beta-glycosidase LXYL-P1-2 reveals the product binding state of GH3 family and a specific pocket for Taxol recognition.
Commun Biol, 3, 2020
|
|
2OQW
 
 | | The crystal structure of sortase B from B.anthracis in complex with AAEK1 | | Descriptor: | Sortase B | | Authors: | Wu, R, Zhang, R, Marresso, A.W, Schneewind, O, Joachimiak, A. | | Deposit date: | 2007-02-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation of inhibitors by sortase triggers irreversible modification of the active site.
J.Biol.Chem., 282, 2007
|
|
