2TSY
 
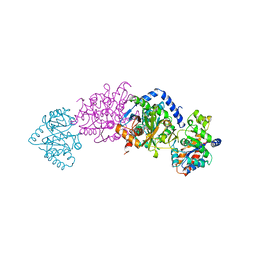 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | SN-GLYCEROL-3-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE, ... | | Authors: | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1997-01-03 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
2TYS
 
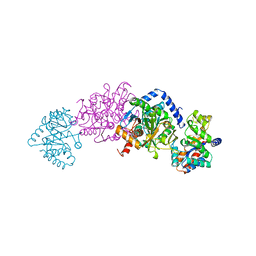 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | SODIUM ION, TRYPTOPHAN SYNTHASE, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-L-TRYPTOPHANE | | Authors: | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1997-01-08 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
6CCX
 
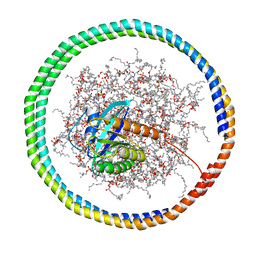 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
1CVO
 
 | |
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
8PHE
 
 | | ACAD9-WT in complex with ECSIT-CTER | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, Evolutionarily conserved signaling intermediate in Toll pathway | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
8PHF
 
 | | Cryo-EM structure of human ACAD9-S191A | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
3VIN
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with a new glucopyranosidic product | | Descriptor: | 2-{4-[2-(beta-D-glucopyranosyloxy)ethyl]piperazin-1-yl}ethanesulfonic acid, Beta-glucosidase, CHLORIDE ION, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3M8J
 
 | |
7HBI
 
 | | SCAPHARCA DIMERIC HEMOGLOBIN, MUTANT T72V, CO-LIGANDED FORM | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Royer Junior, W.E. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mutational destabilization of the critical interface water cluster in Scapharca dimeric hemoglobin: structural basis for altered allosteric activity.
J.Mol.Biol., 284, 1998
|
|
3UWP
 
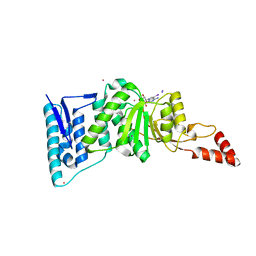 | | Crystal structure of Dot1l in complex with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
6HAW
 
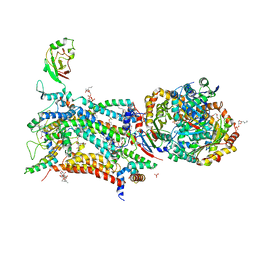 | | Crystal structure of bovine cytochrome bc1 in complex with 2-pyrazolyl quinolone inhibitor WDH2G7 | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ... | | Authors: | Amporndanai, K, Hong, W.D, O'Neill, P.M, Hasnain, S.S, Antonyuk, S.V. | | Deposit date: | 2018-08-08 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Potent Antimalarial 2-Pyrazolyl Quinolonebc1(Qi) Inhibitors with Improved Drug-like Properties.
ACS Med Chem Lett, 9, 2018
|
|
1D4U
 
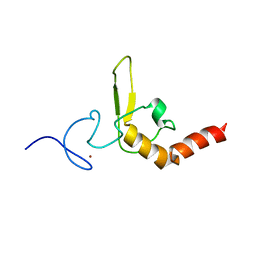 | | INTERACTIONS OF HUMAN NUCLEOTIDE EXCISION REPAIR PROTEIN XPA WITH RPA70 AND DNA: CHEMICAL SHIFT MAPPING AND 15N NMR RELAXATION STUDIES | | Descriptor: | NUCLEOTIDE EXCISION REPAIR PROTEIN XPA (XPA-MBD), ZINC ION | | Authors: | Buchko, G.W, Daughdrill, G.W, de Lorimier, R, Rao, S, Isern, N.G, Lingbeck, J, Taylor, J, Wold, M.S, Gochin, M, Spicer, L.D, Lowry, D.F, Kennedy, M.A. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of human nucleotide excision repair protein XPA with DNA and RPA70 Delta C327: chemical shift mapping and 15N NMR relaxation studies.
Biochemistry, 38, 1999
|
|
3MF7
 
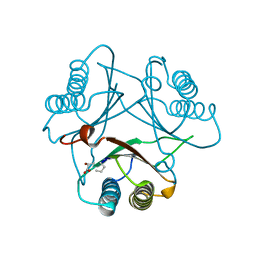 | | Crystal Structure of (R)-oxirane-2-carboxylate inhibited cis-CaaD | | Descriptor: | Cis-3-chloroacrylic acid dehalogenase | | Authors: | Guo, Y, Serrano, H, Ernst, S.R, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of native and inactivated cis-3-chloroacrylic acid dehalogenase: Implications for the catalytic and inactivation mechanisms.
Bioorg.Chem., 39, 2011
|
|
8A0K
 
 | | crystal structure of the kinetoplastid kinetochore protein Trypanosoma brucei KKT3 Divergent Polo-Box domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protein kinase, putative | | Authors: | Ishii, M, Ludzia, P, Marciano, G, Allen, W, Nerusheva, O.O, Akiyoshi, B. | | Deposit date: | 2022-05-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Divergent polo boxes in KKT2 bind KKT1 to initiate the kinetochore assembly cascade in Trypanosoma brucei.
Mol.Biol.Cell, 33, 2022
|
|
6D9Z
 
 | | Structure of CysZ, a sulfate permease from Pseudomonas Denitrificans | | Descriptor: | Sulfate transporter CysZ, octyl beta-D-glucopyranoside | | Authors: | Sanghai, Z.A, Clarke, O.B, Liu, Q, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Mancia, F. | | Deposit date: | 2018-04-30 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4021318 Å) | | Cite: | Structure-based analysis of CysZ-mediated cellular uptake of sulfate.
Elife, 7, 2018
|
|
8HTU
 
 | | Cryo-EM structure of PpPSI-L | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Li, M, Pan, X.W, Sun, H.Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insights into the assembly and energy transfer of the Lhcb9-dependent photosystem I from moss Physcomitrium patens.
Nat.Plants, 9, 2023
|
|
4NHB
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio desulfuricans (Ddes_1525), Target EFI-510107, with bound sn-glycerol-3-phosphate | | Descriptor: | IODIDE ION, SN-GLYCEROL-3-PHOSPHATE, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-04 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
2UWI
 
 | | Structure of CrmE, a poxvirus TNF receptor | | Descriptor: | CRME PROTEIN | | Authors: | Graham, S.C, Bahar, M.W, Abrescia, N.G, Smith, G.L, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2007-03-22 | | Release date: | 2007-07-10 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Crme, a Virus-Encoded Tumour Necrosis Factor Receptor.
J.Mol.Biol., 372, 2007
|
|
6HQS
 
 | | Crystal structure of GcoA F169S bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4NF0
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM PSEUDOMONAS AERUGINOSA PAO1 (PA4616), TARGET EFI-510182, WITH BOUND L-Malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Probable c4-dicarboxylate-binding protein, SULFATE ION | | Authors: | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-30 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NL1
 
 | |
7E2I
 
 | | Cryo-EM structure of hDisp1NNN-ShhN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1, ... | | Authors: | Li, W, Wang, L, Gong, X. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural insights into proteolytic activation of the human Dispatched1 transporter for Hedgehog morphogen release.
Nat Commun, 12, 2021
|
|
8I0A
 
 | |
7E2G
 
 | | Cryo-EM structure of hDisp1NNN-3C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1,Protein dispatched homolog 1 | | Authors: | Li, W, Wang, L, Gong, X. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structural insights into proteolytic activation of the human Dispatched1 transporter for Hedgehog morphogen release.
Nat Commun, 12, 2021
|
|
