5N90
 
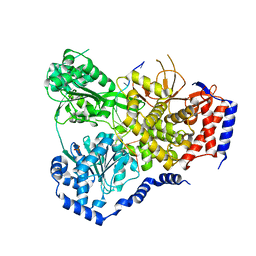 | | Crystal Structure of Drosophila DHX36 helicase in complex with TTGTGGTGT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*TP*TP*GP*TP*GP*GP*TP*GP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.069 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5JCA
 
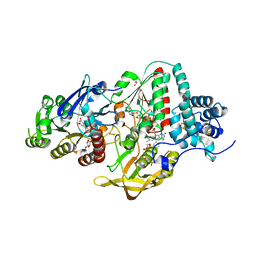 | | NADP(H) bound NADH-dependent Ferredoxin:NADP Oxidoreductase (NfnI) from Pyrococcus furiosus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Zadvornyy, O.A, Schut, G.J, Nguyen, D.M, Artz, J.H, Tokmina-Lukaszewska, M, Lipscomb, G, Adams, M.W, Peters, J.W. | | Deposit date: | 2016-04-14 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic insights into energy conservation by flavin-based electron bifurcation.
Nat. Chem. Biol., 13, 2017
|
|
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
1TBR
 
 | |
1TBQ
 
 | |
5N8S
 
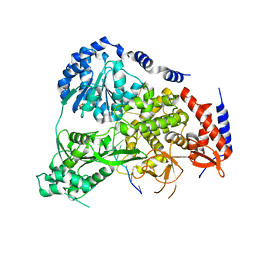 | | Crystal Structure of Drosophila DHX36 helicase in complex with polyT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5CRO
 
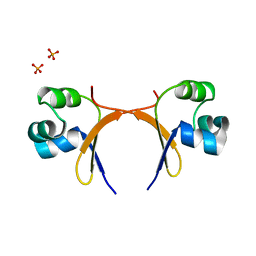 | |
5N96
 
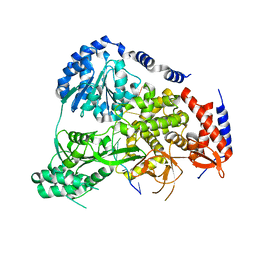 | | Crystal Structure of Drosophila DHX36 helicase in complex with AGGGTTTTTT | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*AP*GP*GP*GP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.716 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N8R
 
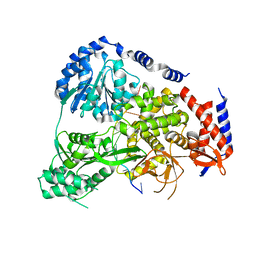 | | Crystal Structure of Drosophilia DHX36 helicase in complex with GAGCACTGC | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*GP*AP*GP*CP*AP*CP*TP*GP*C)-3') | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
3AT1
 
 | |
6TMN
 
 | | Structures of two thermolysin-inhibitor complexes that differ by a single hydrogen bond | | Descriptor: | CALCIUM ION, N-[(2R,4S)-4-hydroxy-2-(2-methylpropyl)-4-oxido-7-oxo-9-phenyl-3,8-dioxa-6-aza-4-phosphanonan-1-oyl]-L-leucine, THERMOLYSIN, ... | | Authors: | Tronrud, D.E, Holden, H.M, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two thermolysin-inhibitor complexes that differ by a single hydrogen bond.
Science, 235, 1987
|
|
1CDN
 
 | |
1BWF
 
 | | ESCHERICHIA COLI GLYCEROL KINASE MUTANT WITH BOUND ATP ANALOG SHOWING SUBSTANTIAL DOMAIN MOTION | | Descriptor: | GLYCEROL, GLYCEROL KINASE, MAGNESIUM ION, ... | | Authors: | Bystrom, C.E, Pettigrew, D.W, Branchaud, B.P, Remington, S.J. | | Deposit date: | 1998-09-23 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Escherichia coli glycerol kinase variant S58-->W in complex with nonhydrolyzable ATP analogues reveal a putative active conformation of the enzyme as a result of domain motion.
Biochemistry, 38, 1999
|
|
6NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
1CDL
 
 | |
1BPM
 
 | |
1CLK
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES DIASTATICUS NO.7 STRAIN M1033 XYLOSE ISOMERASE AT 1.9 A RESOLUTION WITH PSEUDO-I222 SPACE GROUP | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Niu, L, Teng, M, Zhu, X, Gong, W. | | Deposit date: | 1999-04-29 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of xylose isomerase from Streptomyces diastaticus no. 7 strain M1033 at 1.85 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1CXH
 
 | | COMPLEX OF CGTASE WITH MALTOTETRAOSE AT ROOM TEMPERATURE AND PH 9.6 BASED ON DIFFRACTION DATA OF A CRYSTAL SOAKED WITH MALTOHEPTAOSE | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Knegtel, R.M.A, Strokopytov, B.V, Dijkstra, B.W. | | Deposit date: | 1995-07-31 | | Release date: | 1995-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products.
J.Biol.Chem., 270, 1995
|
|
7NWK
 
 | | Crystal structure of CDK9-Cyclin T1 bound by compound 6 | | Descriptor: | Cyclin-T1, Cyclin-dependent kinase 9, N-((1R,3R)-3-(7-(4-fluoro-2-methoxyphenyl)-3H-imidazo[4,5-b]pyridin-2-yl)cyclopentyl)acetamide | | Authors: | Collie, G.W, Ferguson, A.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Series of 7-Azaindoles as Potent and Highly Selective CDK9 Inhibitors for Transient Target Engagement.
J.Med.Chem., 64, 2021
|
|
8RA0
 
 | | Crystal structure of CysF | | Descriptor: | AMP-dependent synthetase, CITRATE ANION, TETRAETHYLENE GLYCOL | | Authors: | Levy, C.W. | | Deposit date: | 2023-11-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Cryptic enzymatic assembly of peptides armed with beta-lactone warheads.
Nat.Chem.Biol., 20, 2024
|
|
1TGT
 
 | | ON THE DISORDERED ACTIVATION DOMAIN IN TRYPSINOGEN. CHEMICAL LABELLING AND LOW-TEMPERATURE CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, METHANOL, TRYPSINOGEN | | Authors: | Walter, J, Steigemann, W, Singh, T.P, Bartunik, H, Bode, W, Huber, R. | | Deposit date: | 1981-10-26 | | Release date: | 1982-03-04 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | On the Disordered Activation Domain in Trypsinogen. Chemical Labelling and Low-Temperature Crystallography
Acta Crystallogr.,Sect.B, 38, 1982
|
|
1CXE
 
 | | COMPLEX OF CGTASE WITH MALTOTETRAOSE AT ROOM TEMPERATURE AND PH 9.1 BASED ON DIFFRACTION DATA OF A CRYSTAL SOAKED WITH ALPHA-CYCLODEXTRIN | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Knegtel, R.M.A, Strokopytov, B.V, Dijkstra, B.W. | | Deposit date: | 1995-07-28 | | Release date: | 1995-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products.
J.Biol.Chem., 270, 1995
|
|
1TRQ
 
 | |
8FG2
 
 | | SARS-CoV-2 Nucleocapsid dimer structure determined from COVID-19 patients | | Descriptor: | Nucleoprotein | | Authors: | Casasanta, M, Jonaid, G.M, Kaylor, L, Luqiu, W, DiCecco, L, Solares, M, Berry, S, Kelly, D.F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural Insights of the SARS-CoV-2 Nucleocapsid Protein: Implications for the Inner-workings of Rapid Antigen Tests.
Microsc Microanal, 29, 2023
|
|
1CZ8
 
 | | VASCULAR ENDOTHELIAL GROWTH FACTOR IN COMPLEX WITH AN AFFINITY MATURED ANTIBODY | | Descriptor: | HEAVY CHAIN OF NEUTRALIZING ANTIBODY, LIGHT CHAIN OF NEUTRALIZING ANTIBODY, SULFATE ION, ... | | Authors: | Chen, Y, Wiesmann, C, Fuh, G, Li, B, Christinger, H.W, McKay, P, de Vos, A.M, Lowman, H.B. | | Deposit date: | 1999-09-01 | | Release date: | 2000-03-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selection and analysis of an optimized anti-VEGF antibody: crystal structure of an affinity-matured Fab in complex with antigen.
J.Mol.Biol., 293, 1999
|
|
