4W8R
 
 | |
6Q5H
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24D | | Descriptor: | AMMONIUM ION, CC-Hex*-L24D, SULFATE ION | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5K
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24K | | Descriptor: | AMMONIUM ION, CC-Hex*-L24K, GLYCEROL, ... | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
4WB5
 
 | | Crystal structure of human cAMP-dependent protein kinase A (catalytic alpha subunit) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PKI (5-24), ... | | Authors: | Cheung, J, Ginter, C, Cassidy, M, Franklin, M.C, Rudolph, M.J, Hendrickson, W.A. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural insights into mis-regulation of protein kinase A in human tumors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6PU5
 
 | | MicroED structure of proteinase K recorded on CetaD | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Hattne, J, Martynowycz, M.W, Penzcek, P.A, Gonen, T. | | Deposit date: | 2019-07-17 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.7 Å) | | Cite: | MicroED with the Falcon III direct electron detector.
Iucrj, 6, 2019
|
|
6PUR
 
 | | Human TRPM2 bound to ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
6PWR
 
 | |
4WD3
 
 | | Crystal structure of an L-amino acid ligase RizA | | Descriptor: | L-amino acid ligase | | Authors: | Kagawa, W, Arai, T, Kino, K, Kurumizaka, H. | | Deposit date: | 2014-09-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of RizA, an L-amino-acid ligase from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6PVX
 
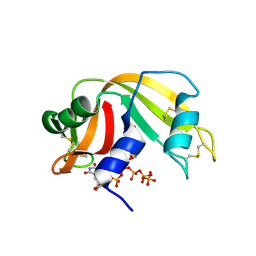 | | RNase A in complex with p5U | | Descriptor: | 5'-O-[(R)-hydroxy{[(S)-hydroxy{[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]oxy}phosphoryl]uridine, Ribonuclease pancreatic | | Authors: | Windsor, I.W, Sheppard, S.M, Cummins, C.C, Raines, R.T. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Nucleoside Tetra- and Pentaphosphates Prepared Using a Tetraphosphorylation Reagent Are Potent Inhibitors of Ribonuclease A.
J.Am.Chem.Soc., 141, 2019
|
|
6PZ9
 
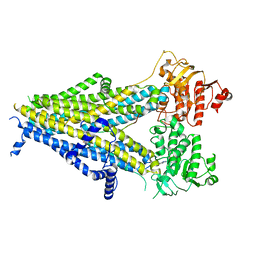 | | Cryo-EM structure of the pancreatic beta-cell SUR1 bound to ATP and repaglinide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ATP-sensitive inward rectifier potassium channel 11, ... | | Authors: | Shyng, S.L, Yoshioka, C, Martin, G.M, Sung, M.W. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Mechanism of pharmacochaperoning in a mammalian K ATP channel revealed by cryo-EM.
Elife, 8, 2019
|
|
1TMN
 
 | |
6QAQ
 
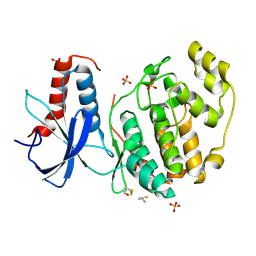 | | ERK2 mini-fragment binding | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, thiophen-3-ylmethylazanium | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
6Q1M
 
 | | Crystal structure of the wheat dwarf virus Rep domain | | Descriptor: | GLYCEROL, Replication-associated protein | | Authors: | Litzau, L.A, Everett, B.A, Evans III, R.L, Shi, K, Tompkins, K, Gordon, W.R. | | Deposit date: | 2019-08-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of the Wheat dwarf virus Rep domain.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6Q3D
 
 | | Dye type peroxidase Aa from Streptomyces lividans: 235.2 kGy structure | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | Deposit date: | 2018-12-04 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
4W5Z
 
 | |
6Q5M
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24Dab | | Descriptor: | CC-Hex*-L24Dab, SODIUM ION | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q7V
 
 | |
6Q8F
 
 | | Nterminal domain of human SMU1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, WD40 repeat-containing protein SMU1 | | Authors: | Tengo, L, Le Corre, L, Fournier, G, Ashraf, U, Busca, P, Rameix-Welti, M.-A, Gravier-Pelletier, C, Ruigrok, R.W.H, Jacob, Y, Vidalain, P.-O, Pietrancosta, N, Naffakh, N, McCarthy, A.A, Crepin, T. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Destabilization of the human RED-SMU1 splicing complex as a basis for host-directed antiinfluenza strategy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6Q8K
 
 | | CLK1 with bound pyridoquinazoline | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase CLK1, ~{N}2-(3-morpholin-4-ylpropyl)pyrido[3,4-g]quinazoline-2,10-diamine | | Authors: | Schroeder, M, Tazarki, H, Zeinyeh, W, Esvan, Y.J, Khiari, J, Joesselin, B, Bach, S, Ruchaud, S, Anizon, F, Giraud, F, Moreau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis.
Eur J Med Chem, 166, 2019
|
|
6QAL
 
 | | ERK2 mini-fragment binding | | Descriptor: | 1,1-bis(oxidanylidene)thietan-3-ol, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
6QAW
 
 | | ERK2 mini-fragment binding | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, [1-(7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-yl]methylazanium | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
4UWO
 
 | | Native di-zinc VIM-26. Leu224 in VIM-26 from Klebsiella pneumoniae has implications for drug binding. | | Descriptor: | GLYCEROL, METALLO-BETA-LACTAMASE VIM-26, SODIUM ION, ... | | Authors: | Leiros, H.-K.S, Edvardsen, K.S.W, Bjerga, G.E.K, Samuelsen, O. | | Deposit date: | 2014-08-14 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.555 Å) | | Cite: | Structural and Biochemical Characterization of Vim-26 Show that Leu224 Has Implications for the Substrate Specificity of Vim Metallo-Beta-Lactamases.
FEBS J., 282, 2015
|
|
6Q25
 
 | |
4UWR
 
 | | Mono-zinc VIM-26. Leu224 in VIM-26 from Klebsiella pneumoniae has implications for drug binding. | | Descriptor: | FORMIC ACID, METALLO-BETA-LACTAMASE VIM-26, SODIUM ION, ... | | Authors: | Leiros, H.-K.S, Edvardsen, K.S.W, Bjerga, G.E.K, Samuelsen, O. | | Deposit date: | 2014-08-14 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Biochemical Characterization of Vim-26 Show that Leu224 Has Implications for the Substrate Specificity of Vim Metallo-Beta-Lactamases.
FEBS J., 282, 2015
|
|
6Q3E
 
 | | Dye type peroxidase Aa from Streptomyces lividans: 274.4 kGy structure | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | Deposit date: | 2018-12-04 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
