8D1N
 
 | | bBest2_345 Ca2+-bound open state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1F
 
 | | hBest2 5mM Ca2+ (Ca2+-bound) closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.82 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
3A4Z
 
 | | Structure of cytochrome P450 Vdh mutant (Vdh-K1) obtained by directed evolution | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
6V0T
 
 | | Crystal Structure of Catalytic Subunit of Bovine Pyruvate Dehydrogenase Phosphatase 1 - Catalytic Domain | | Descriptor: | MANGANESE (II) ION, SULFATE ION, [Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 1, ... | | Authors: | Guo, Y, Qiu, W, Ernst, S.R, Carroll, D.W, Hackert, M.L. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the catalytic subunit of bovine pyruvate dehydrogenase phosphatase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6F88
 
 | |
8D1E
 
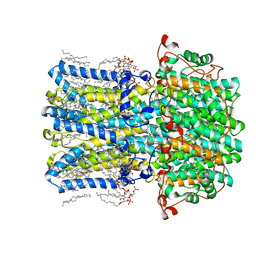 | | hBest2 1uM Ca2+ (Ca2+-bound) closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.78 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
4QIA
 
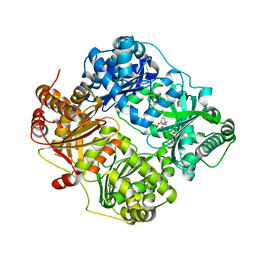 | | Crystal structure of human insulin degrading enzyme (ide) in complex with inhibitor N-benzyl-N-(carboxymethyl)glycyl-L-histidine | | Descriptor: | Insulin-degrading enzyme, N-benzyl-N-(carboxymethyl)glycyl-L-histidine, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structure-activity relationships of imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, dual binders of human insulin-degrading enzyme.
Eur.J.Med.Chem., 90, 2015
|
|
6V2H
 
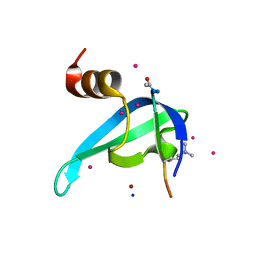 | | Crystal structure of CDYL2 in complex with H3tK27me3 | | Descriptor: | Chromodomain Y-like protein 2, H3tK27me3, NICKEL (II) ION, ... | | Authors: | Dong, C, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
5I7I
 
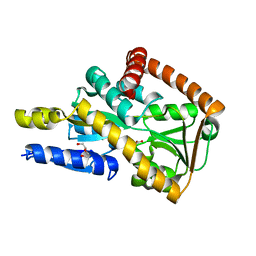 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-crystallized 3-hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, DI(HYDROXYETHYL)ETHER, TRAP solute Binding Protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-02-17 | | Release date: | 2017-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-purified 4-hydroxybenzoate
To Be Published
|
|
8D1L
 
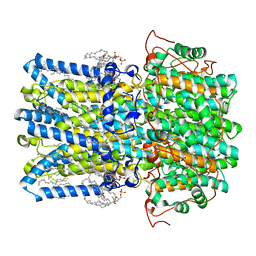 | | hBest1 Ca2+-bound partially open aperture state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
3S74
 
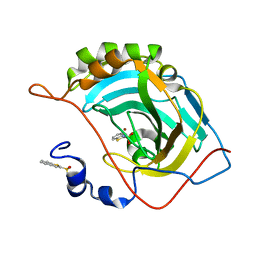 | |
5O5P
 
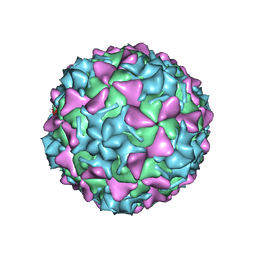 | | Poliovirus type 3 (strain Saukett) stabilized virus-like particle in complex with the pocket factor compound GPP3 | | Descriptor: | 1-[5-[4-(ethoxyiminomethyl)phenoxy]-3-methyl-pentyl]-3-pyridin-4-yl-imidazol-2-one, Capsid proteins, VP4, ... | | Authors: | Bahar, M.W, Kotecha, A, Fry, E.E, Stuart, D.I. | | Deposit date: | 2017-06-02 | | Release date: | 2017-07-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Plant-made polio type 3 stabilized VLPs-a candidate synthetic polio vaccine.
Nat Commun, 8, 2017
|
|
7EHT
 
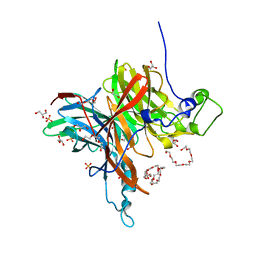 | | Levansucrase from Brenneria sp. EniD 312 | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, GLYCEROL, Levansucrase, ... | | Authors: | Xu, W, Hou, X.D, Rao, Y.J, Pijning, T, Guskov, A, Mu, W.M. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram-Negative Bacterium Brenneria Provides Insights into Its Product Size Specificity.
J.Agric.Food Chem., 70, 2022
|
|
8D1J
 
 | | hBest1 5mM Ca2+ (Ca2+-bound) closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
6OZJ
 
 | |
5I0C
 
 | | Crystal structure of predicted acyltransferase YjdJ with acyl-CoA N-acyltransferase domain from Escherichia coli str. K-12 | | Descriptor: | CADMIUM ION, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-03 | | Release date: | 2016-02-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of predicted acyltransferase YjdJ with acyl-CoA N-acyltransferase domain from Escherichia coli str. K-12
To Be Published
|
|
6OZQ
 
 | |
5IE3
 
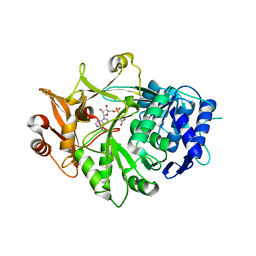 | | Crystal structure of a plant enzyme | | Descriptor: | ADENOSINE MONOPHOSPHATE, OXALIC ACID, Oxalate--CoA ligase | | Authors: | Fan, M.R, Li, M, Chang, W.R. | | Deposit date: | 2016-02-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana Oxalyl-CoA Synthetase Essential for Oxalate Degradation
Mol Plant, 9, 2016
|
|
8HUD
 
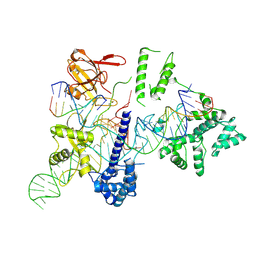 | | Cryo-EM structure of the EvCas9-sgRNA-target DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target DNA strand, Target DNA strand, ... | | Authors: | Tang, N, Wu, Z, Gao, Y, Chen, W, Su, M, Wang, Z, Ji, Q. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Molecular Basis and Genome Editing Applications of a Compact Eubacterium ventriosum CRISPR-Cas9 System.
Acs Synth Biol, 13, 2024
|
|
6ET2
 
 | |
8DJF
 
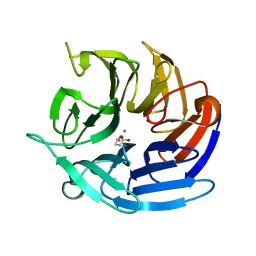 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound tetrahedral intermediate | | Descriptor: | (5S)-5-methyloxolane-2,2-diol, CALCIUM ION, Gluconolactonase, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2022-06-30 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
8DK0
 
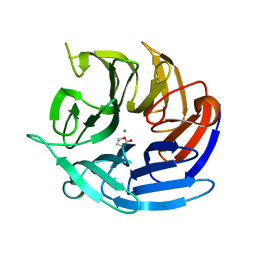 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound (S)gamma-valerolactone | | Descriptor: | CALCIUM ION, Gluconolactonase, SODIUM ION, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
3S75
 
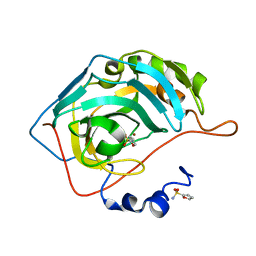 | |
5WAP
 
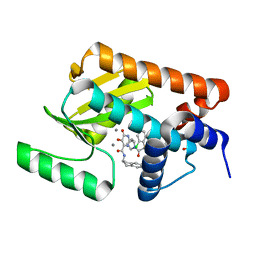 | | Crystal structure of the influenza virus PA endonuclease in complex with inhibitor 10i (SRI-30025) | | Descriptor: | 2-[(2S)-1-{[(2-chlorophenyl)sulfanyl]acetyl}pyrrolidin-2-yl]-N-(2,3-dihydro-1H-inden-2-yl)-5-hydroxy-6-oxo-1,6-dihydropyrimidine-4-carboxamide, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-06-26 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
5I7E
 
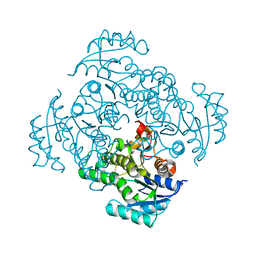 | |
