1FEH
 
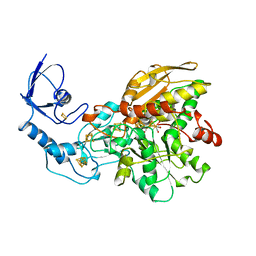 | | FE-ONLY HYDROGENASE FROM CLOSTRIDIUM PASTEURIANUM | | Descriptor: | 2 IRON/2 SULFUR/5 CARBONYL/2 WATER INORGANIC CLUSTER, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Peters, J.W, Lanzilotta, W.N, Lemon, B.J, Seefeldt, L.C. | | Deposit date: | 1998-10-28 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution.
Science, 282, 1998
|
|
8TL0
 
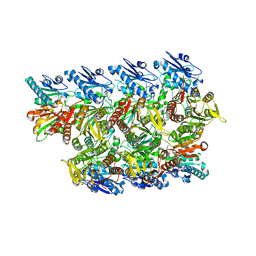 | |
3K9Z
 
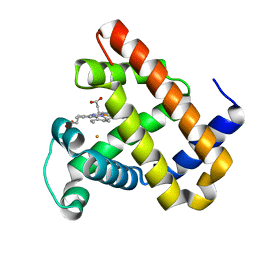 | | Rational Design of a Structural and Functional Nitric Oxide Reductase | | Descriptor: | FE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yeung, N, Lin, Y.-W, Gao, Y.-G, Zhao, X, Russell, B.S, Lei, L, Miner, K.D, Robinson, H, Lu, Y. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of a structural and functional nitric oxide reductase.
Nature, 462, 2009
|
|
2YIA
 
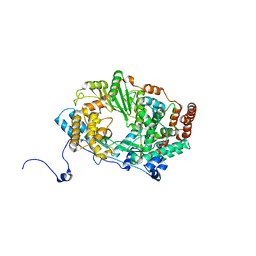 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus | | Descriptor: | POTASSIUM ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
7QQJ
 
 | | Sucrose phosphorylase from Jeotgalibaca ciconiae | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Sucrose phosphorylase | | Authors: | Ubiparip, Z, Capra, N, Rozeboom, H.J, Desmet, T, Thunnissen, A.M.W.H. | | Deposit date: | 2022-01-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Sucrose phosphorylase from Jeotgalibaca ciconiae
To Be Published
|
|
2YJT
 
 | |
1FK3
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH PALMITOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, PALMITOLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
3NE6
 
 | | RB69 DNA Polymerase (S565G/Y567A) Ternary Complex with dCTP Opposite dG | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-06-08 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Variation in Mutation Rates Caused by RB69pol Fidelity Mutants Can Be Rationalized on the Basis of Their Kinetic Behavior and Crystal Structures.
J.Mol.Biol., 406, 2011
|
|
1FGQ
 
 | |
3NGI
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dTTP Opposite dG | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), DNA (5'-D(*TP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), ... | | Authors: | Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-06-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.886 Å) | | Cite: | Variation in Mutation Rates Caused by RB69pol Fidelity Mutants Can Be Rationalized on the Basis of Their Kinetic Behavior and Crystal Structures.
J.Mol.Biol., 406, 2011
|
|
7QQI
 
 | | Sucrose phosphorylase from Faecalibaculum rodentium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aamy domain-containing protein | | Authors: | Ubiparip, Z, Capra, N, Rozeboom, H.J, Desmet, T, Thunnissen, A.M.W.H. | | Deposit date: | 2022-01-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Sucrose phosphorylase from Faecalibaculum rodentium
To Be Published
|
|
1FSB
 
 | | STRUCTURE OF THE EGF DOMAIN OF P-SELECTIN, NMR, 19 STRUCTURES | | Descriptor: | P-SELECTIN | | Authors: | Freedman, S.J, Sanford, D.G, Bachovchin, W.W, Furie, B.C, Baleja, J.D, Furie, B. | | Deposit date: | 1996-03-25 | | Release date: | 1997-04-01 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the epidermal growth factor domain of P-selectin.
Biochemistry, 35, 1996
|
|
4ZJ0
 
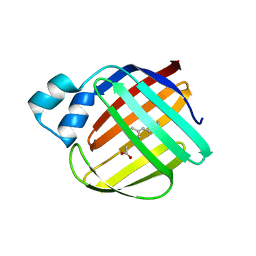 | | The crystal structure of monomer Q108K:K40L:Y60W CRBPII bound to all-trans-retinal | | Descriptor: | ACETATE ION, RETINAL, Retinol-binding protein 2 | | Authors: | Nossoni, Z, Assar, Z, Wang, W, Vasileiou, C, Borhan, B, Geiger, J.H. | | Deposit date: | 2015-04-28 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Domain-Swapped Dimers of Intracellular Lipid-Binding Proteins: Evidence for Ordered Folding Intermediates.
Structure, 24, 2016
|
|
1PJG
 
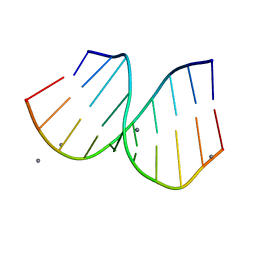 | | RNA/DNA Hybrid Decamer of CAAAGAAAAG/CTTTTCTTTG | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', CALCIUM ION | | Authors: | Kopka, M.L, Lavelle, L, Han, G.W, Ng, H.-L, Dickerson, R.E. | | Deposit date: | 2003-06-02 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | An unusual sugar conformation in the structure of an RNA/DNA decamer of the polypurine tract may affect recognition by RNase H.
J.Mol.Biol., 334, 2003
|
|
4BTL
 
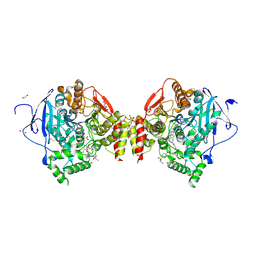 | | Aromatic interactions in acetylcholinesterase-inhibitor complexes | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Qian, W, Engdahl, C, Berg, L, Ekstrom, F, Linusson, A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3JD8
 
 | | cryo-EM structure of the full-length human NPC1 at 4.4 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Gong, X, Qian, H.W, Zhou, X.H, Wu, J.P, Zhou, Q, Yan, N. | | Deposit date: | 2016-05-01 | | Release date: | 2016-06-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | The structure of Niemann-Pick C1 protein
To Be Published
|
|
8EHX
 
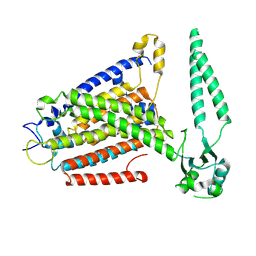 | | cryo-EM structure of TMEM63B in LMNG | | Descriptor: | CSC1-like protein 2 | | Authors: | Zheng, W, Fu, T.M, Holt, J.R. | | Deposit date: | 2022-09-14 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | TMEM63 proteins function as monomeric high-threshold mechanosensitive ion channels.
Neuron, 111, 2023
|
|
1JA3
 
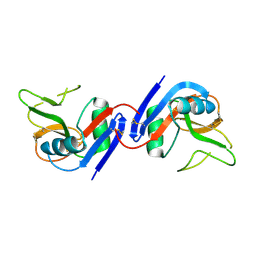 | | Crystal Structure of the Murine NK Cell Inhibitory Receptor Ly-49I | | Descriptor: | MHC class I recognition receptor Ly49I | | Authors: | Dimasi, N, Sawicki, W.M, Reineck, L.A, Li, Y, Natarajan, K, Murgulies, D.H, Mariuzza, A.R. | | Deposit date: | 2001-05-29 | | Release date: | 2002-07-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Ly49I natural killer cell receptor reveals variability in dimerization mode within the Ly49 family.
J.Mol.Biol., 320, 2002
|
|
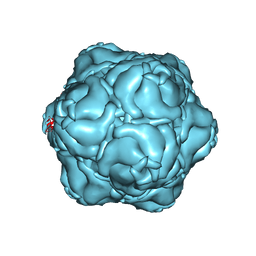
8T6R
 
 | |
7QWC
 
 | |
5BWZ
 
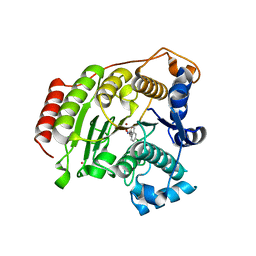 | |
6VR6
 
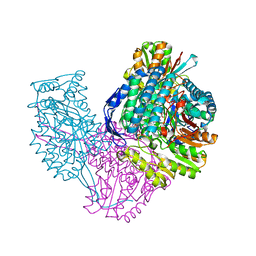 | | Structure of ALDH9A1 complexed with NAD+ in space group P1 | | Descriptor: | 4-trimethylaminobutyraldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wyatt, J.W, Tanner, J.J. | | Deposit date: | 2020-02-06 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch.Biochem.Biophys., 691, 2020
|
|
5BXY
 
 | | Crystal structure of RNA methyltransferase from Salinibacter ruber in complex with S-Adenosyl-L-homocysteine | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA methyltransferase, ... | | Authors: | Handing, K.B, LaRowe, C, Shabalin, I.G, Stead, M, Hillerich, B.S, Ahmed, M, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-01 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of RNA methylase family protein from Salinibacterruber in complex with S-Adenosyl-L-homocysteine.
to be published
|
|
5BYW
 
 | |
1PJO
 
 | | Crystal Structure of an RNA/DNA hybrid of HIV-1 PPT | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', MAGNESIUM ION | | Authors: | Kopka, M.L, Lavelle, L, Han, G.W, Ng, H.-L, Dickerson, R.E. | | Deposit date: | 2003-06-03 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An Unusual Sugar Conformation in the Structure of an RNA/DNA
Decamer of the Polypurine Tract May Affect Recognition by RNase H
J.Mol.Biol., 334, 2003
|
|
