1Z3Z
 
 | | The crystal structure of a DGD mutant: Q52A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Fogle, E.J, Liu, W, Toney, M.D. | | Deposit date: | 2005-03-14 | | Release date: | 2006-01-03 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Role of q52 in catalysis of decarboxylation and transamination in dialkylglycine decarboxylase.
Biochemistry, 44, 2005
|
|
7PZT
 
 | | Structure of the bacterial toxin, TecA, an asparagine deamidase from Alcaligenes faecalis. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Urea amidohydrolase | | Authors: | Dix, S.R, Aziz, A.A, Baker, P.J, Evans, C.A, Dickman, M.J, Farthing, R.J, King, Z.L.S, Nathan, S, Partridge, L.J, Raih, F.M, Sedelnikova, S.E, Thomas, M.S, Rice, D.W. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structure of A. faecalis TecA provides insights into its role as an asparagine deamidase toxin which targets RhoA
To Be Published
|
|
2E6B
 
 | |
5M0M
 
 | | Structure-based evolution of a hybrid steroid series of Autotaxin inhibitors | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Keune, W.-J, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2016-10-05 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Design of Autotaxin Inhibitors by Structural Evolution of Endogenous Modulators.
J. Med. Chem., 60, 2017
|
|
5M0S
 
 | | Structure-based evolution of a hybrid steroid series of Autotaxin inhibitors | | Descriptor: | CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, GLYCEROL, ... | | Authors: | Keune, W.-J, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2016-10-05 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Design of Autotaxin Inhibitors by Structural Evolution of Endogenous Modulators.
J. Med. Chem., 60, 2017
|
|
3JXQ
 
 | | X-Ray structure of r[CGCG(5-fluoro)CG]2 | | Descriptor: | MAGNESIUM ION, r[CGCG(5-fluoro)CG]2 | | Authors: | Adamiak, D.A, Milecki, J, Adamiak, R.W, Rypniewski, W. | | Deposit date: | 2009-09-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The hydration and unusual hydrogen bonding in the crystal structure of an RNA duplex containing alternating CG base pairs
New J.Chem., 34, 2010
|
|
5ET1
 
 | | Crystal structure of Myo3b-ARB1 in complex with Espin1-AR | | Descriptor: | Espin, GLYCEROL, Myosin-IIIb | | Authors: | Liu, H, Li, J, liu, W, Zhang, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Myosin III-mediated cross-linking and stimulation of actin bundling activity of Espin
Elife, 5, 2016
|
|
3D81
 
 | | Sir2-S-alkylamidate complex crystal structure | | Descriptor: | NAD-dependent deacetylase, S-alkylamidate intermediate, ZINC ION | | Authors: | Hawse, W.F, Hoff, K.G, Fatkins, D, Daines, A, Zubkova, O.V, Schramm, V.L, Zheng, W, Wolberger, C. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into intermediate steps in the Sir2 deacetylation reaction.
Structure, 16, 2008
|
|
1RAB
 
 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
1KW5
 
 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-01-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
5M8S
 
 | | Crystal structure of human tyrosinase related protein 1 mutant (T391V-R374S-Y362F) in complex with phenylthiourea (PTU) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, X, Soler-Lopez, M, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2016-10-29 | | Release date: | 2018-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phenylthiourea Binding to Human Tyrosinase-Related Protein 1
Int J Mol Sci, 2020
|
|
5MD5
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=11, twist=0 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MDD
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=23, twist=6 | | Descriptor: | Gag protein | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MD1
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=5, twist=0 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
3HXB
 
 | | Engineered RabGGTase in complex with a peptidomimetic inhibitor (compound 6) | | Descriptor: | Geranylgeranyl transferase type-2 subunit alpha, Geranylgeranyl transferase type-2 subunit beta, N-[(benzyloxy)carbonyl]-D-tyrosyl-L-phenylalanyl-L-tyrosine, ... | | Authors: | Guo, Z, Alexandrov, K, Waldmann, H, Goody, R.S, Blankenfeldt, W. | | Deposit date: | 2009-06-20 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design, synthesis, and characterization of Peptide-based rab geranylgeranyl transferase inhibitors
J.Med.Chem., 52, 2009
|
|
6RDT
 
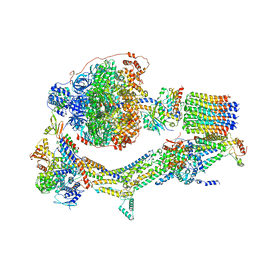 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1E, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
5FSR
 
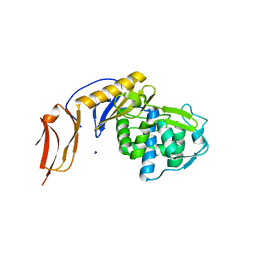 | | Crystal structure of penicillin binding protein 6B from Escherichia coli | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE DACD | | Authors: | Peters, K, Kannan, S, Rao, V.A, Bilboy, J, Vollmer, D, Erickson, S.W, Lewis, R.J, Young, K.D, Vollmer, W. | | Deposit date: | 2016-01-07 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Redundancy of Peptidoglycan Carboxypeptidases Ensures Robust Cell Shape Maintenance in Escherichia Coli
Mbio, 7, 2016
|
|
6REA
 
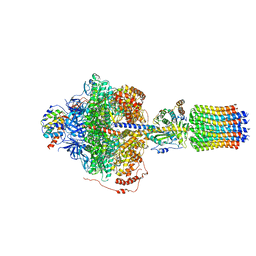 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2D, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RES
 
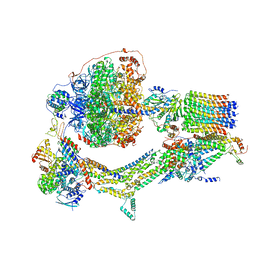 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3C, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
5MHD
 
 | |
2AJ8
 
 | | Porcine dipeptidyl peptidase IV (CD26) in complex with 7-Benzyl-1,3-dimethyl-8-piperazin-1-yl-3,7-dihydro-purine-2,6-dione (BDPX) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-BENZYL-1,3-DIMETHYL-8-PIPERAZIN-1-YL-3,7-DIHYDRO-PURINE-2,6-DIONE, ... | | Authors: | Engel, M, Hoffmann, T, Manhart, S, Heiser, U, Chambre, S, Huber, R, Demuth, H.U, Bode, W. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
J.Mol.Biol., 355, 2006
|
|
6U6O
 
 | | Crystal structure of a vaccine-elicited anti-HIV-1 rhesus macaque antibody DH846 | | Descriptor: | DH846 Fab heavy chain, DH846 Fab light chain | | Authors: | Chen, W.-H, Choe, M, Saunders, K.O, Joyce, M.G. | | Deposit date: | 2019-08-30 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structural and genetic convergence of HIV-1 neutralizing antibodies in vaccinated non-human primates.
Plos Pathog., 17, 2021
|
|
5FII
 
 | | Structure of a human aspartate kinase, chorismate mutase and TyrA domain. | | Descriptor: | PHENYLALANINE, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Patel, D, Kopec, J, Shrestha, L, Fitzpatrick, F, Pinkas, D, Chaikuad, A, Dixon-Clarke, S, McCorvie, T.J, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2015-09-25 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Ligand-Dependent Dimerization of Phenylalanine Hydroxylase Regulatory Domain.
Sci.Rep., 6, 2016
|
|
3FGA
 
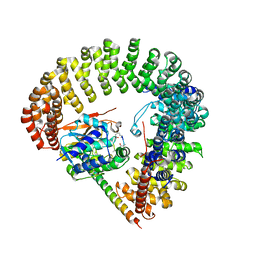 | | Structural Basis of PP2A and Sgo interaction | | Descriptor: | MANGANESE (II) ION, MICROCYSTIN-LR, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | Authors: | Xu, Z, Xu, W. | | Deposit date: | 2008-12-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of the PP2A-shugoshin interaction
Mol.Cell, 35, 2009
|
|
6RD5
 
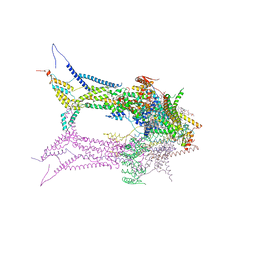 | | CryoEM structure of Polytomella F-ATP synthase, focussed refinement of Fo and peripheral stalk, C2 symmetry | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ATP synthase associated protein ASA1, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
