6M7P
 
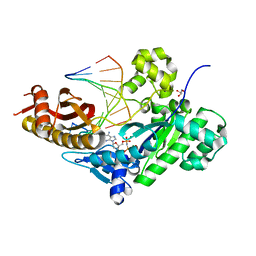 | | Human DNA polymerase eta extension complex with cdA at the -2 position | | 分子名称: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*TP*GP*TP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*TP*CP*(02I)P*CP*AP*CP*AP*CP*T)-3'), ... | | 著者 | Gregory, M.T, Yang, W. | | 登録日 | 2018-08-20 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Bypassing a 8,5'-cyclo-2'-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3L6G
 
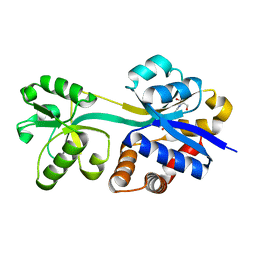 | | Crystal structure of lactococcal OpuAC in its open conformation | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Betaine ABC transporter permease and substrate binding protein | | 著者 | Berntsson, R.P.A, Wolters, J.C, Gul, N, Karasawa, A, Thunnissen, A.M.W.H, Slotboom, D.J, Poolman, B. | | 登録日 | 2009-12-23 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Ligand binding and crystal structures of the substrate-binding domain of the ABC transporter OpuA.
Plos One, 5, 2010
|
|
7NAB
 
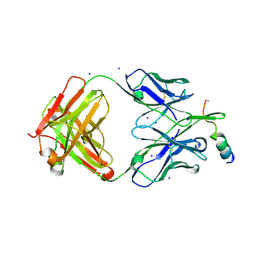 | | Crystal structure of human neutralizing mAb CV3-25 binding to SARS-CoV-2 S MPER peptide 1140-1165 | | 分子名称: | CITRIC ACID, CV3-25 Fab Heavy Chain, CV3-25 Fab Light Chain, ... | | 著者 | Chen, Y, Tolbert, W.D, Pazgier, M. | | 登録日 | 2021-06-21 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern.
Cell Rep, 38, 2022
|
|
6X6U
 
 | |
8AT1
 
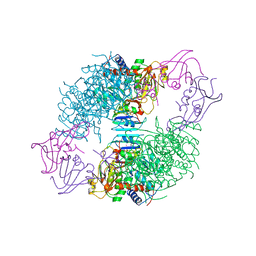 | | CRYSTAL STRUCTURES OF ASPARTATE CARBAMOYLTRANSFERASE LIGATED WITH PHOSPHONOACETAMIDE, MALONATE, AND CTP OR ATP AT 2.8-ANGSTROMS RESOLUTION AND NEUTRAL P*H | | 分子名称: | ASPARTATE CARBAMOYLTRANSFERASE (R STATE), CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ... | | 著者 | Gouaux, J.E, Stevens, R.C, Lipscomb, W.N. | | 登録日 | 1989-09-22 | | 公開日 | 1990-10-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of aspartate carbamoyltransferase ligated with phosphonoacetamide, malonate, and CTP or ATP at 2.8-A resolution and neutral pH.
Biochemistry, 29, 1990
|
|
8ATC
 
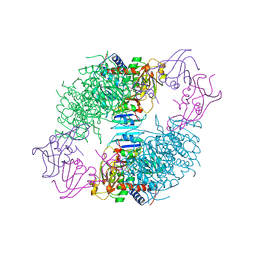 | | COMPLEX OF N-PHOSPHONACETYL-L-ASPARTATE WITH ASPARTATE CARBAMOYLTRANSFERASE. X-RAY REFINEMENT, ANALYSIS OF CONFORMATIONAL CHANGES AND CATALYTIC AND ALLOSTERIC MECHANISMS | | 分子名称: | ASPARTATE CARBAMOYLTRANSFERASE (R STATE), CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ... | | 著者 | Ke, H, Lipscomb, W.N, Cho, Y, Honzatko, R.B. | | 登録日 | 1989-08-25 | | 公開日 | 1990-10-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Complex of N-phosphonacetyl-L-aspartate with aspartate carbamoyltransferase. X-ray refinement, analysis of conformational changes and catalytic and allosteric mechanisms.
J.Mol.Biol., 204, 1988
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | 分子名称: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-04-14 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | 分子名称: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-04-28 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7QKB
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound GC376 | | 分子名称: | CHLORIDE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | 登録日 | 2021-12-17 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
6ZFH
 
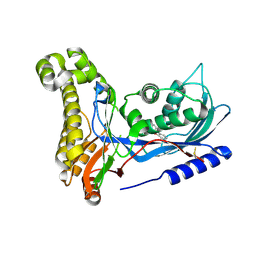 | | Structure of human galactokinase in complex with galactose and 2'-(benzo[d]oxazol-2-ylamino)-7',8'-dihydro-1'H-spiro[cyclopentane-1,4'-quinazolin]-5'(6'H)-one | | 分子名称: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclopentane]-5-one, Galactokinase, beta-D-galactopyranose | | 著者 | Bezerra, G.A, Mackinnon, S, Zhang, M, Foster, W, Bailey, H, Arrowsmith, C, Edwards, A, Bountra, C, Lai, K, Yue, W.W, Structural Genomics Consortium (SGC) | | 登録日 | 2020-06-17 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.439 Å) | | 主引用文献 | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
6ZH0
 
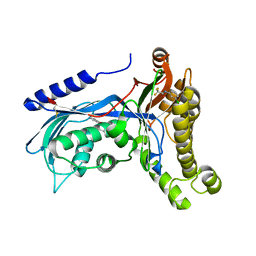 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | 分子名称: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(3-chlorophenyl)-2,2,2-trifluoroacetamide, ... | | 著者 | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | 登録日 | 2020-06-20 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | 分子名称: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | 著者 | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-09-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
5MWV
 
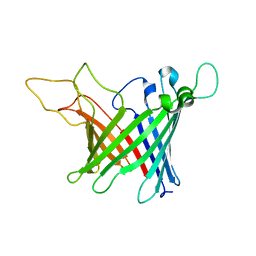 | | Solid-state NMR Structure of outer membrane protein G in lipid bilayers | | 分子名称: | Outer membrane protein G | | 著者 | Retel, J.S, Nieuwkoop, A.J, Hiller, M, Higman, V.A, Barbet-Massin, E, Stanek, J, Andreas, L.B, Franks, W.T, van Rossum, B.-J, Vinothkumar, K.R, Handel, L, de Palma, G.G, Bardiaux, B, Pintacuda, G, Emsley, L, Kuelbrandt, W, Oschkinat, H. | | 登録日 | 2017-01-20 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structure of outer membrane protein G in lipid bilayers.
Nat Commun, 8, 2017
|
|
7RIK
 
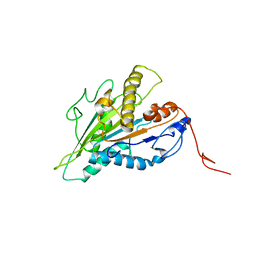 | | Magic-Angle-Spinning NMR Structure of Kinesin-1 Motor Domain Assembled with Microtubules | | 分子名称: | Kinesin-1 heavy chain | | 著者 | Zhang, C, Guo, C, Russell, R.W, Quinn, C.M, Li, M, Williams, J.C, Gronenborn, A.M, Polenova, T. | | 登録日 | 2021-07-20 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Magic-angle-spinning NMR structure of the kinesin-1 motor domain assembled with microtubules reveals the elusive neck linker orientation
Nat Commun, 13, 2022
|
|
7D2C
 
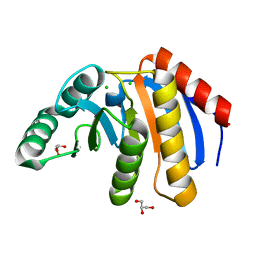 | | The Crystal Structure of human PARP14 from Biortus. | | 分子名称: | CHLORIDE ION, GLYCEROL, Protein mono-ADP-ribosyltransferase PARP14 | | 著者 | Wang, F, Miao, Q, Lv, Z, Cheng, W, Lin, D, Xu, X, Tan, J. | | 登録日 | 2020-09-16 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | The Crystal Structure of human PARP14 from Biortus.
To Be Published
|
|
7D4F
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to suramin | | 分子名称: | 8-(3-(3-aminobenzamido)-4-methylbenzamido)naphthalene-1,3,5-trisulfonic acid, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Li, Z, Yin, W, Zhou, Z, Yu, X, Xu, H. | | 登録日 | 2020-09-23 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D7P
 
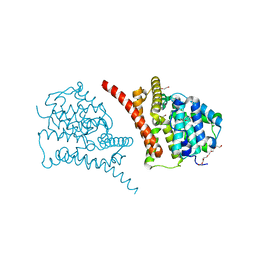 | | Crystal structure of the phosphodiesterase domain of Salpingoeca rosetta rhodopsin phosphodiesterase | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | 著者 | Ikuta, T, Shihoya, W, Yamashita, K, Nureki, O. | | 登録日 | 2020-10-05 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into the mechanism of rhodopsin phosphodiesterase.
Nat Commun, 11, 2020
|
|
7DFV
 
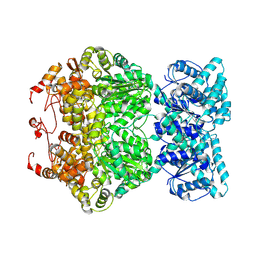 | | Cryo-EM structure of plant NLR RPP1 tetramer core part | | 分子名称: | NAD+ hydrolase (NADase) | | 著者 | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | 登録日 | 2020-11-10 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7D20
 
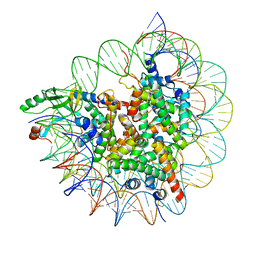 | | Cryo-EM structure of SET8-CENP-A-nucleosome complex | | 分子名称: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Ho, C.-H, Takizawa, Y, Kobayashi, W, Arimura, Y, Kurumizaka, H. | | 登録日 | 2020-09-15 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase.
Life Sci Alliance, 4, 2021
|
|
7D1Z
 
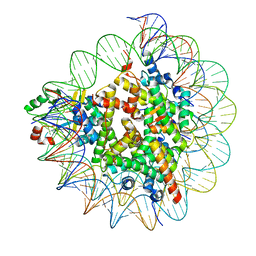 | | Cryo-EM structure of SET8-nucleosome complex | | 分子名称: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Ho, C.-H, Takizawa, Y, Kobayashi, W, Arimura, Y, Kurumizaka, H. | | 登録日 | 2020-09-15 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase.
Life Sci Alliance, 4, 2021
|
|
7DCL
 
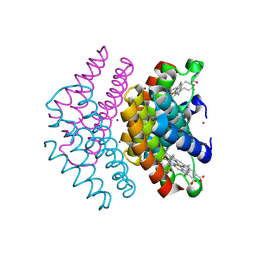 | |
7D1G
 
 | | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase GAPDH from Clostridium beijerinckii | | 分子名称: | BETA-MERCAPTOETHANOL, Glyceraldehyde-3-phosphate dehydrogenase, MAGNESIUM ION | | 著者 | Chen, Y, Lan, J, Liu, W, Wang, L, Xu, Y. | | 登録日 | 2020-09-14 | | 公開日 | 2021-03-17 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase GAPDH from Clostridium beijerinckii
To Be Published
|
|
7N9F
 
 | | Structure of the in situ yeast NPC | | 分子名称: | Dynein light chain 1, cytoplasmic, Nucleoporin 145c, ... | | 著者 | Villa, E, Singh, D, Ludtke, S.J, Akey, C.W, Rout, M.P, Echeverria, I, Suslov, S. | | 登録日 | 2021-06-17 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (37 Å) | | 主引用文献 | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7DKG
 
 | | Influenza H5N1 nucleoprotein (truncated) in complex with nucleotides | | 分子名称: | Nucleoprotein, RNA (5'-R(P*(OMU)P*(OMU)P*(OMU))-3') | | 著者 | Tang, Y.S, Xu, S, Chen, Y.W, Wang, J.H, Shaw, P.C. | | 登録日 | 2020-11-24 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structures of influenza nucleoprotein complexed with nucleic acid provide insights into the mechanism of RNA interaction.
Nucleic Acids Res., 49, 2021
|
|
7DDA
 
 | | Envelope protein VP37 a crystal structure from White Spot Syndrome Virus | | 分子名称: | Envelope protein, SULFATE ION | | 著者 | Somsoros, W, Sangawa, T, Takebe, K, Attarataya, J, Suzuki, M, Khunrae, P. | | 登録日 | 2020-10-28 | | 公開日 | 2021-06-23 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Crystal structure of the C-terminal domain of envelope protein VP37 from white spot syndrome virus reveals sulphate binding sites responsible for heparin binding.
J.Gen.Virol., 102, 2021
|
|
