8SFQ
 
 | |
7KRV
 
 | |
7KRT
 
 | |
3NR2
 
 | | Crystal structure of Caspase-6 zymogen | | 分子名称: | Caspase-6 | | 著者 | Su, X.-D, Wang, X.-J, Liu, X, Mi, W, Wang, K.-T. | | 登録日 | 2010-06-30 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation
Embo Rep., 11, 2010
|
|
5C5I
 
 | | Crystal structure of NADP-dependent dehydrogenase from Rhodobacter sphaeroides | | 分子名称: | NADP-dependent dehydrogenase | | 著者 | Kowiel, M, Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Porebski, P.J, Cymborowski, M, Al Obaidi, N.F, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2015-06-19 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of NADP-dependent dehydrogenase from Rhodobacter sphaeroides
to be published
|
|
5TA3
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 2) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | 著者 | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | 登録日 | 2016-09-09 | | 公開日 | 2016-10-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
8SFN
 
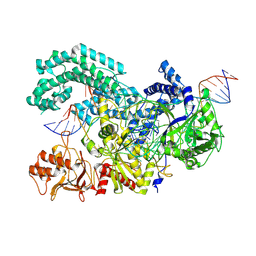 | |
8SFI
 
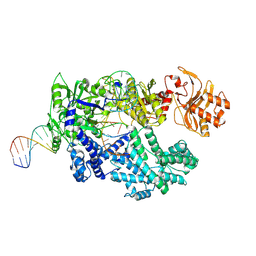 | | WT CRISPR-Cas12a with a 8bp R-loop | | 分子名称: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*CP*TP*TP*AP*TP*CP*AP*CP*TP*AP*AP*AP*AP*GP*AP*TP*CP*GP*GP*AP*AP*G)-3'), DNA (5'-D(P*CP*TP*TP*CP*CP*GP*AP*TP*CP*TP*TP*TP*TP*AP*GP*TP*GP*AP*TP*A)-3'), ... | | 著者 | Strohkendl, I, Taylor, D.W. | | 登録日 | 2023-04-11 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | WT CRISPR-Cas12a with a 8bp R-loop
To Be Published
|
|
8SFP
 
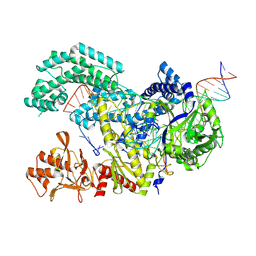 | |
8SFH
 
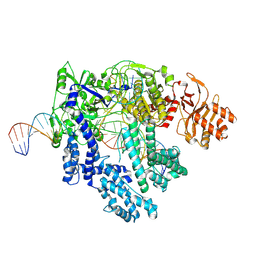 | |
5C5S
 
 | |
8SFR
 
 | |
8FO4
 
 | |
4I41
 
 | | Crystal Structure of human Ser/Thr kinase Pim1 in complex with mitoxantrone | | 分子名称: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, Serine/threonine-protein kinase pim-1 | | 著者 | Zhang, W, Wan, X, Xie, Y, Huang, N. | | 登録日 | 2012-11-27 | | 公開日 | 2013-12-11 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A new target for an old drug: identifying mitoxantrone as a nanomolar inhibitor of PIM1 kinase via kinome-wide selectivity modeling.
J. Med. Chem., 56, 2013
|
|
1DWF
 
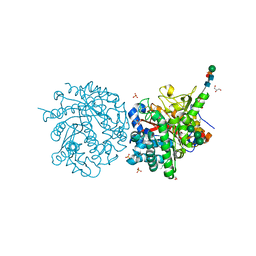 | |
2XZE
 
 | |
3M2R
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | 分子名称: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, Coenzyme B, ... | | 著者 | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | 登録日 | 2010-03-08 | | 公開日 | 2010-09-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
2XF0
 
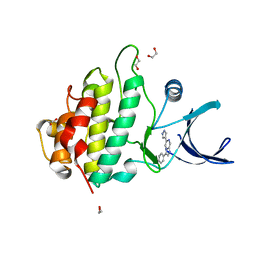 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | 分子名称: | 1,2-ETHANEDIOL, 3-PHENYL-6-(1H-PYRAZOL-4-YL)IMIDAZO[1,2-A]PYRAZINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | 著者 | Matthews, T.P, McHardy, T, Klair, S, Boxall, K, Fisher, M, Cherry, M, Allen, C.E, Addison, G.J, Ellard, J, Aherne, G.W, Westwood, I.M, van Montfort, R, Garrett, M.D, Reader, J.C, Collins, I. | | 登録日 | 2010-05-19 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Design and Evaluation of 3,6-Di(Hetero)Aryl Imidazo[1,2-A]Pyrazines as Inhibitors of Checkpoint and Other Kinases.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3M9K
 
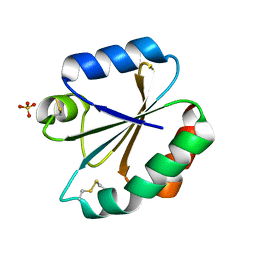 | | Crystal structure of human thioredoxin C69/73S double-mutant, oxidized form | | 分子名称: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, SULFATE ION, Thioredoxin | | 著者 | Weichsel, A, Montfort, W.R. | | 登録日 | 2010-03-22 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of human thioredoxin revealing an unraveled helix and exposed S-nitrosation site.
Protein Sci., 19, 2010
|
|
5BVT
 
 | | Palmitate-bound pFABP5 | | 分子名称: | Epidermal fatty acid-binding protein, PALMITOLEIC ACID | | 著者 | Lee, J.H, Lee, C.W, Do, H. | | 登録日 | 2015-06-05 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
3IR4
 
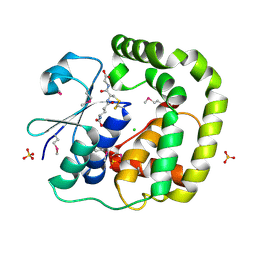 | | 1.2 Angstrom Crystal Structure of the Glutaredoxin 2 (grxB) from Salmonella typhimurium in complex with Glutathione | | 分子名称: | CHLORIDE ION, GLUTATHIONE, Glutaredoxin 2, ... | | 著者 | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Halavaty, A, Dauter, Z, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-08-21 | | 公開日 | 2009-09-01 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | 1.2 Angstrom Crystal Structure of the Glutaredoxin 2 (grxB) from Salmonella typhimurium in complex with Glutathione.
TO BE PUBLISHED
|
|
6GG4
 
 | | Crystal structure of M2 PYK in complex with Phenyalanine. | | 分子名称: | PHENYLALANINE, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | McNae, I.W, Yuan, M, Walkinshaw, M.D. | | 登録日 | 2018-05-02 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | An allostatic mechanism for M2 pyruvate kinase as an amino-acid sensor.
Biochem. J., 475, 2018
|
|
5BVS
 
 | | Linoleate-bound pFABP4 | | 分子名称: | Fatty acid-binding protein, LINOLEIC ACID | | 著者 | Lee, J.H, Lee, C.W, Do, H. | | 登録日 | 2015-06-05 | | 公開日 | 2015-08-05 | | 最終更新日 | 2015-09-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
6VH3
 
 | | 2.20 A resolution structure of MERS 3CL protease in complex with inhibitor 7j | | 分子名称: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | 著者 | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-01-09 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
4E2F
 
 | |
