2MNY
 
 | | NMR Structure of KDM5B PHD1 finger | | 分子名称: | Lysine-specific demethylase 5B, ZINC ION | | 著者 | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | 登録日 | 2014-04-16 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
4WKU
 
 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | 分子名称: | Acyl-homoserine lactone acylase PvdQ, GLYCEROL, hexyl(trihydroxy)borate(1-) | | 著者 | Wu, R, Clevenger, K.D, Fast, W, Liu, D. | | 登録日 | 2014-10-03 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
6LVP
 
 | |
2M04
 
 | |
1AQ6
 
 | | STRUCTURE OF L-2-HALOACID DEHALOGENASE FROM XANTHOBACTER AUTOTROPHICUS | | 分子名称: | FORMIC ACID, L-2-HALOACID DEHALOGENASE | | 著者 | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | 登録日 | 1997-08-07 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Three-dimensional structure of L-2-haloacid dehalogenase from Xanthobacter autotrophicus GJ10 complexed with the substrate-analogue formate.
J.Biol.Chem., 272, 1997
|
|
7AS5
 
 | | 126 helix bundle DNA nanostructure | | 分子名称: | SCAFFOLD STRAND, STAPLE STRAND | | 著者 | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | 登録日 | 2020-10-27 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (9.8 Å) | | 主引用文献 | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
1RJP
 
 | | Crystal structure of D-aminoacylase in complex with 100mM CuCl2 | | 分子名称: | ACETATE ION, COPPER (II) ION, D-aminoacylase, ... | | 著者 | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | 登録日 | 2003-11-20 | | 公開日 | 2004-04-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
8A8M
 
 | | Structure of the MAPK p38alpha in complex with its activating MAP2K MKK6 | | 分子名称: | Dual specificity mitogen-activated protein kinase kinase 6, MAGNESIUM ION, Mitogen-activated protein kinase 14, ... | | 著者 | Bowler, M.W, Juyoux, P, Pellegrini, E. | | 登録日 | 2022-06-23 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Architecture of the MKK6-p38 alpha complex defines the basis of MAPK specificity and activation.
Science, 381, 2023
|
|
3VBF
 
 | | Crystal structure of formaldehyde treated human Enterovirus 71 (space group I23) | | 分子名称: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Genome Polyprotein, ... | | 著者 | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | 登録日 | 2012-01-02 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7ART
 
 | | 48 helix bundle DNA origami brick | | 分子名称: | SCAFFOLD STRAND, STAPLE STRAND | | 著者 | Feigl, E, Kube, M, Kohler, F, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | 登録日 | 2020-10-26 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
4WT7
 
 | | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Agrobacterium vitis (Avi_5165, Target EFI-511223) with bound allitol | | 分子名称: | ABC transporter substrate binding protein (Ribose), CHLORIDE ION, D-allitol | | 著者 | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-10-29 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Agrobacterium vitis (Avi_5165, Target EFI-511223) with bound allitol
To be published
|
|
5YDE
 
 | | Crystal structure of a disease-related gene, hCDC73(1-111) | | 分子名称: | Parafibromin | | 著者 | Sun, W, Kuang, X.L, Liu, Y.P, Tian, L.F, Yan, X.X, Xu, W.Q. | | 登録日 | 2017-09-13 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.023 Å) | | 主引用文献 | Crystal structure of the N-terminal domain of human CDC73 and its implications for the hyperparathyroidism-jaw tumor (HPT-JT) syndrome
Sci Rep, 7, 2017
|
|
6EHJ
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and peptide bound | | 分子名称: | ASPARAGINE, COENZYME A, GLYCEROL, ... | | 著者 | Perez-Dorado, I, Ritzefeld, M, Tate, E.W. | | 登録日 | 2017-09-13 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
8CPA
 
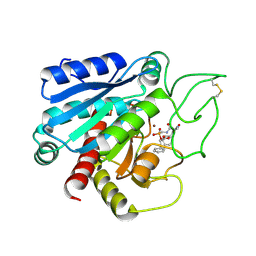 | |
2HI3
 
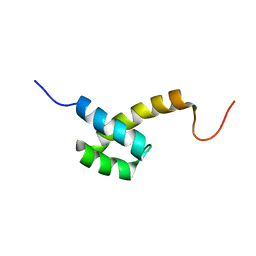 | | Solution structure of the homeodomain-only protein HOP | | 分子名称: | Homeodomain-only protein | | 著者 | Mackay, J.P, Kook, H, Epstein, J.A, Simpson, R.J, Yung, W.W. | | 登録日 | 2006-06-28 | | 公開日 | 2007-01-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Analysis of the structure and function of the transcriptional coregulator HOP
Biochemistry, 45, 2006
|
|
3VIK
 
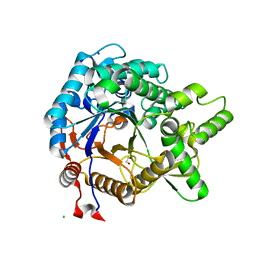 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with cellobiose | | 分子名称: | 1,2-ETHANEDIOL, Beta-glucosidase, CHLORIDE ION, ... | | 著者 | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | 登録日 | 2011-10-03 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VJ9
 
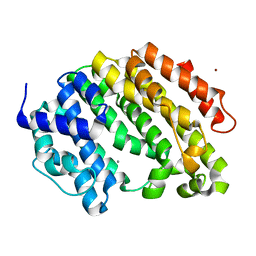 | | Crystal structure of the human squalene synthase | | 分子名称: | CALCIUM ION, NICKEL (II) ION, Squalene synthase | | 著者 | Liu, C.I, Jeng, W.Y, Chang, W.J, Wang, A.H.J. | | 登録日 | 2011-10-14 | | 公開日 | 2012-04-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Binding modes of zaragozic acid A to human squalene synthase and staphylococcal dehydrosqualene synthase
J.Biol.Chem., 287, 2012
|
|
6J4N
 
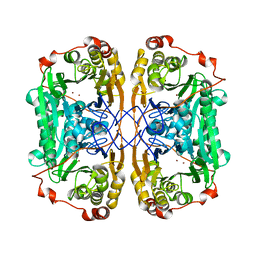 | | Structure of papua new guinea MBL-1(PNGM-1) native | | 分子名称: | Metallo-beta-lactamases PNGM-1, ZINC ION | | 著者 | Hong, M.K, Park, K.S, Jeon, J.H, Lee, J.H, Park, Y.S, Lee, S.H, Kang, L.W. | | 登録日 | 2019-01-10 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The novel metallo-beta-lactamase PNGM-1 from a deep-sea sediment metagenome: crystallization and X-ray crystallographic analysis.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6LXE
 
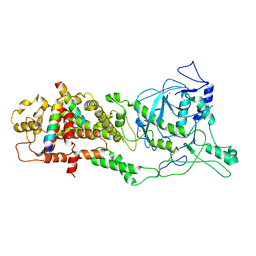 | | DROSHA-DGCR8 complex | | 分子名称: | Microprocessor complex subunit DGCR8, Ribonuclease 3, ZINC ION | | 著者 | Jin, W, Wang, J, Liu, C.P, Wang, H.W, Xu, R.M. | | 登録日 | 2020-02-10 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural Basis for pri-miRNA Recognition by Drosha.
Mol.Cell, 78, 2020
|
|
4X7G
 
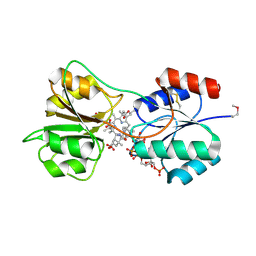 | | CobK precorrin-6A reductase | | 分子名称: | 3-[(1R,2S,3S,7S,11S,16S,17R,18R,19R)-2,7,12,18-tetrakis(2-hydroxy-2-oxoethyl)-3,13-bis(3-hydroxy-3-oxopropyl)-1,2,5,7,11,17-hexamethyl-17-(3-oxidanyl-3-oxidanylidene-prop-1-enyl)-3,6,8,10,15,16,18,19,21,24-decahydrocorrin-8-yl]propanoic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Precorrin-6A reductase | | 著者 | Gu, S, Deery, E, Warren, M.J, Pickersgill, R.W. | | 登録日 | 2014-12-09 | | 公開日 | 2016-01-20 | | 最終更新日 | 2017-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Crystal structure of CobK reveals strand-swapping between Rossmann-fold domains and molecular basis of the reduced precorrin product trap.
Sci Rep, 5, 2015
|
|
7TB3
 
 | | cryo-EM structure of MBP-KIX-apoferritin | | 分子名称: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed | | 著者 | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | 登録日 | 2021-12-21 | | 公開日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
4WJI
 
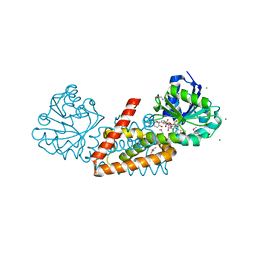 | | Crystal structure of cyclohexadienyl dehydrogenase from Sinorhizobium meliloti in complex with NADP and tyrosine | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Shabalin, I.G, Cooper, D.R, Hou, J, Zimmerman, M.D, Stead, M, Hillerich, B.S, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-09-30 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of cyclohexadienyl dehydrogenase from Sinorhizobium meliloti in complex with NADP
to be published
|
|
2RFB
 
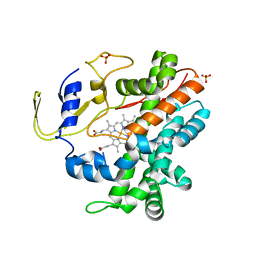 | | Crystal Structure of a Cytochrome P450 from the Thermoacidophilic Archaeon Picrophilus Torridus | | 分子名称: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | 著者 | Ho, W.W, Li, H, Poulos, T.L, Nishida, C.R, Ortiz de Montellano, P.R. | | 登録日 | 2007-09-28 | | 公開日 | 2008-01-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure and Properties of CYP231A2 from the Thermoacidophilic Archaeon Picrophilus torridus.
Biochemistry, 47, 2008
|
|
6LTR
 
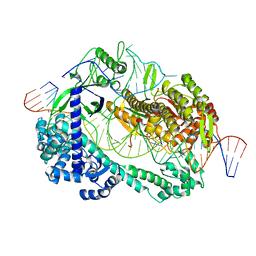 | | Crystal structure of Cas12i2 ternary complex with single Mg2+ bound in catalytic pocket | | 分子名称: | 1,2-ETHANEDIOL, Cas12i2, DNA (35-MER), ... | | 著者 | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | 登録日 | 2020-01-23 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
7T4O
 
 | |
