7RVW
 
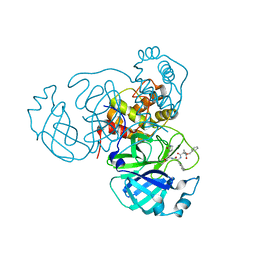 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI23 | | 分子名称: | 3C-like proteinase, benzyl (1-{[(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamoyl}cyclopropyl)carbamate | | 著者 | Yang, K, Liu, W. | | 登録日 | 2021-08-19 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
3VCY
 
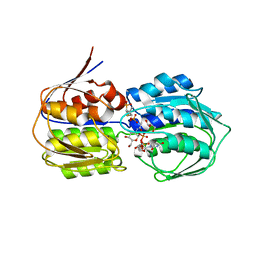 | | Structure of MurA (UDP-N-acetylglucosamine enolpyruvyl transferase), from Vibrio fischeri in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin. | | 分子名称: | GLYCEROL, PHOSPHATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | 著者 | Bensen, D.C, Rodriguez, S, Nix, J, Cunningham, M.L, Tari, L.W. | | 登録日 | 2012-01-04 | | 公開日 | 2012-04-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.925 Å) | | 主引用文献 | Structure of MurA (UDP-N-acetylglucosamine enolpyruvyl transferase) from Vibrio fischeri in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7RVU
 
 | |
4DT2
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment CC27209 | | 分子名称: | (2,2-dimethyl-2,3-dihydro-1-benzofuran-7-yl)methanol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Feder, D, Hussein, W.M, Clayton, D.J, Kan, M, Schenk, G, McGeary, R.P, Guddat, L.W. | | 登録日 | 2012-02-20 | | 公開日 | 2012-09-19 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
171L
 
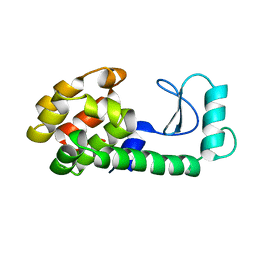 | |
1RJ5
 
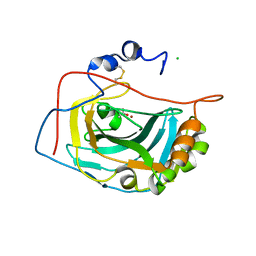 | | Crystal Structure of the Extracellular Domain of Murine Carbonic Anhydrase XIV | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CHLORIDE ION, ... | | 著者 | Whittington, D.A, Grubb, J.H, Waheed, A, Shah, G.N, Sly, W.S, Christianson, D.W. | | 登録日 | 2003-11-18 | | 公開日 | 2004-03-09 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Expression, assay, and structure of the extracellular domain of murine carbonic anhydrase XIV: implications for selective inhibition of membrane-associated isozymes.
J.Biol.Chem., 279, 2004
|
|
7RW1
 
 | |
2FSX
 
 | | Crystal structure of Rv0390 from M. tuberculosis | | 分子名称: | BROMIDE ION, COG0607: Rhodanese-related sulfurtransferase, SULFATE ION | | 著者 | Bursey, E.H, Radhakannan, T, Yu, M, Segelke, B.W, Lekin, T, Toppani, D, Chang, Y.-B, Kaviratne, T, Woodruff, T, Terwilliger, T.C, Hung, L.-W, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2006-01-23 | | 公開日 | 2006-02-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Rv0390 from Mycobacterium tuberculosis
To be Published
|
|
7SGK
 
 | |
7RVZ
 
 | |
7RVV
 
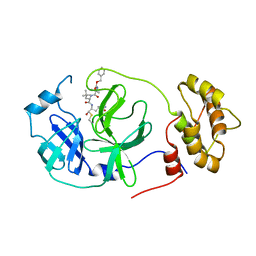 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI22 | | 分子名称: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-2-methyl-L-alanyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | 著者 | Yang, K, Liu, W. | | 登録日 | 2021-08-19 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
2M8S
 
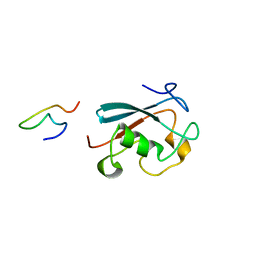 | |
7RVQ
 
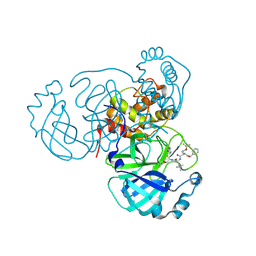 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI16 | | 分子名称: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | 著者 | Yang, K, Liu, W. | | 登録日 | 2021-08-19 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
2MCW
 
 | | Solid-state NMR structure of piscidin 3 in aligned 3:1 phosphatidylcholine/phosphoglycerol lipid bilayers | | 分子名称: | Piscidin-3 | | 著者 | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Pastor, R.W, Cotten, M.L. | | 登録日 | 2013-08-27 | | 公開日 | 2014-01-22 | | 最終更新日 | 2014-03-19 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
3VIF
 
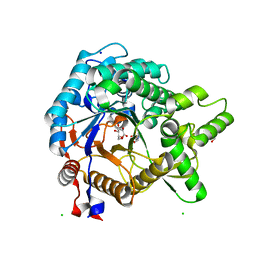 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with gluconolactone | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-glucosidase, CHLORIDE ION, ... | | 著者 | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | 登録日 | 2011-10-03 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7RVX
 
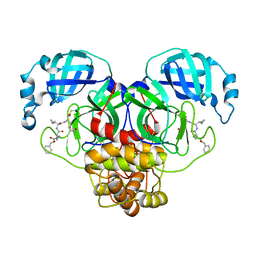 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI24 | | 分子名称: | 3C-like proteinase, benzyl [(1S)-1-cyclopropyl-2-{[(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]amino}-2-oxoethyl]carbamate | | 著者 | Yang, K, Liu, W. | | 登録日 | 2021-08-19 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVT
 
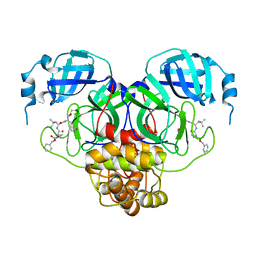 | |
7RVP
 
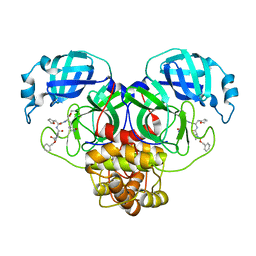 | |
1AC1
 
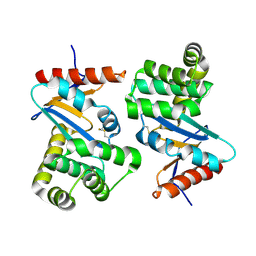 | | DSBA MUTANT H32L | | 分子名称: | DSBA | | 著者 | Guddat, L.W, Martin, J.L. | | 登録日 | 1997-02-10 | | 公開日 | 1997-10-15 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability.
Protein Sci., 6, 1997
|
|
7RVN
 
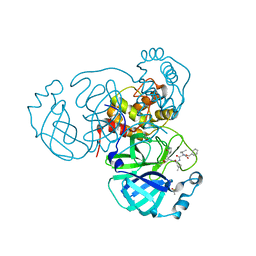 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI12 | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methylidene-L-norvalinamide | | 著者 | Yang, K, Liu, W. | | 登録日 | 2021-08-19 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVY
 
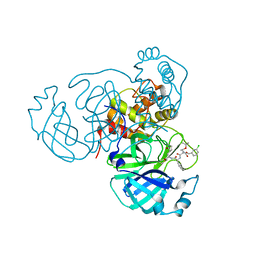 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI25 | | 分子名称: | 3C-like proteinase, O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | 著者 | Yang, K, Liu, W. | | 登録日 | 2021-08-19 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
6O7K
 
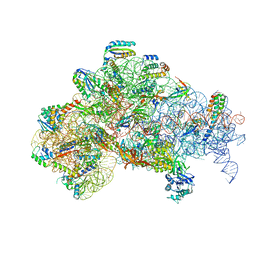 | | 30S initiation complex | | 分子名称: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | 著者 | Frank, J, Gonzalez Jr, R.L, kaledhonkar, S, Fu, Z, Caban, K, Li, W, Chen, B, Sun, M. | | 登録日 | 2019-03-08 | | 公開日 | 2019-05-29 | | 最終更新日 | 2020-01-29 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Late steps in bacterial translation initiation visualized using time-resolved cryo-EM.
Nature, 570, 2019
|
|
6INT
 
 | | xylose isomerase from Paenibacillus sp. R4 | | 分子名称: | CALCIUM ION, Xylose isomerase | | 著者 | Lee, J.H, Lee, C.W, Park, S. | | 登録日 | 2018-10-26 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.942 Å) | | 主引用文献 | Crystal Structure and Functional Characterization of a Xylose Isomerase (PbXI) from the Psychrophilic Soil Microorganism, Paenibacillus sp.
J. Microbiol. Biotechnol., 29, 2019
|
|
1AJM
 
 | |
4WKS
 
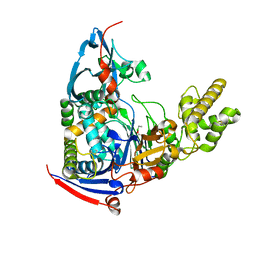 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | 分子名称: | Acyl-homoserine lactone acylase PvdQ, ethylboronic acid | | 著者 | Wu, R, Clevenger, D.K, Fast, W, Liu, D. | | 登録日 | 2014-10-03 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.629 Å) | | 主引用文献 | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
