3QC8
 
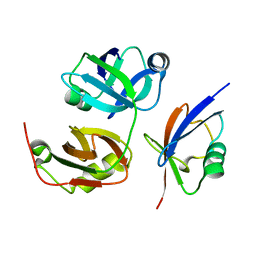 | | Crystal Structure of FAF1 UBX Domain In Complex with p97/VCP N Domain Reveals The Conserved FcisP Touch-Turn Motif of UBX Domain Suffering Conformational Change | | Descriptor: | FAS-associated factor 1, Transitional endoplasmic reticulum ATPase | | Authors: | Kim, K.H, Kang, W, Suh, S.W, Yang, J.K. | | Deposit date: | 2011-01-15 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FAF1 UBX domain in complex with p97/VCP N domain reveals a conformational change in the conserved FcisP touch-turn motif of UBX domain
Proteins, 79, 2011
|
|
6XO1
 
 | | Crystal structure of N97A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.758 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
6XNO
 
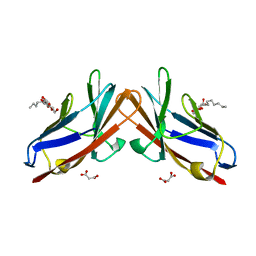 | | Crystal structure of E99A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID, octyl beta-D-glucopyranoside | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-03 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
7N9T
 
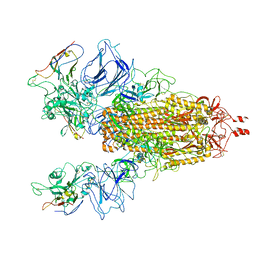 | |
8BBX
 
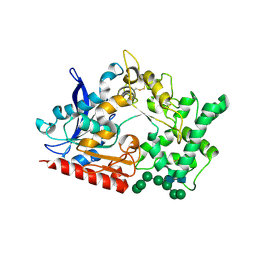 | | Structure of prolyl endoprotease from Aspergillus niger CBS 109712 in space group C222(1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoprotease endo-Pro, TETRAETHYLENE GLYCOL, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Van der Laan, J.M, De Jong, R.M, Dijkstra, B.W. | | Deposit date: | 2022-10-14 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and time-resolved mechanistic investigations of protein hydrolysis by the acidic proline-specific endoprotease from Aspergillus niger.
Protein Sci., 33, 2024
|
|
1SO2
 
 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 3B In COMPLEX WITH A DIHYDROPYRIDAZINE INHIBITOR | | Descriptor: | 1-DEOXY-1-[(2-HYDROXYETHYL)(NONANOYL)AMINO]HEXITOL, 6-(4-{[2-(3-IODOBENZYL)-3-OXOCYCLOHEX-1-EN-1-YL]AMINO}PHENYL)-5-METHYL-4,5-DIHYDROPYRIDAZIN-3(2H)-ONE, MAGNESIUM ION, ... | | Authors: | Scapin, G, Patel, S.B, Chung, C, Varnerin, J.P, Edmondson, S.D, Mastracchio, A, Parmee, E.R, Becker, J.W, Singh, S.B, Van Der Ploeg, L.H, Tota, M.R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Phosphodiesterase 3B: Atomic Basis for Substrate and Inhibitor Specificity
Biochemistry, 43, 2004
|
|
7NP4
 
 | | cAMP-bound rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
6XD4
 
 | | CDC-like protein | | Descriptor: | ACETATE ION, Hemolysin, SODIUM ION | | Authors: | Morton, C.J, Parker, M.W, Lawrence, S.L, Johnstone, B.A, Tweten, R.K. | | Deposit date: | 2020-06-09 | | Release date: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Key Motif in the Cholesterol-Dependent Cytolysins Reveals a Large Family of Related Proteins.
Mbio, 11, 2020
|
|
6XDD
 
 | | Crystal structure of IRE1 in complex with G-3053 | | Descriptor: | 4-[(trans-4-aminocyclohexyl)amino]-N-(6-chloro-3-{[(2,5-difluorophenyl)sulfonyl]amino}-2-fluorophenyl)thieno[3,2-d]pyrimidine-7-carboxamide, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Ackerly-Wallweber, H, Wang, W. | | Deposit date: | 2020-06-10 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of BRaf-Sparing Amino-Thienopyrimidines with Potent IRE1 alpha Inhibitory Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
7NP3
 
 | | cAMP-free rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H.M, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7NFC
 
 | | Cryo-EM structure of NHEJ super-complex (dimer) | | Descriptor: | DNA (27-MER), DNA (28-MER), DNA ligase 4, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
8B57
 
 | | Structure of prolyl endoprotease from Aspergillus niger CBS 109712 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Van der Laan, J.M, De Jong, R.M, Dijkstra, B.W. | | Deposit date: | 2022-09-22 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and time-resolved mechanistic investigations of protein hydrolysis by the acidic proline-specific endoprotease from Aspergillus niger.
Protein Sci., 33, 2024
|
|
7NFE
 
 | | Cryo-EM structure of NHEJ super-complex (monomer) | | Descriptor: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*CP*TP*AP*TP*TP*AP*TP*TP*AP*TP*G)-3'), DNA (5'-D(P*TP*AP*AP*TP*AP*AP*TP*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*AP*G)-3'), DNA ligase 4, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2021-02-06 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
4KAP
 
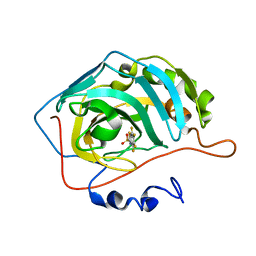 | | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand | | Descriptor: | 4,5,6,7-tetrafluoro-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Lange, H, Lockett, M.R, Breiten, B, Heroux, A, Sherman, W, Rappoport, D, Yau, P.O, Snyder, P.W, Whitesides, G.M. | | Deposit date: | 2013-04-22 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
6XO2
 
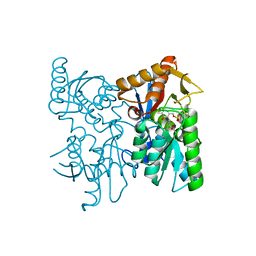 | | Structural Characterization of Beta Cyanoalanine Synthase from Tetranychus Urticae (two-spotted spider mite) | | Descriptor: | ACETATE ION, Beta-cyanoalanine synthase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Daneshian, L, Schlachter, C, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of beta-cyanoalanine synthase from Tetranychus urticae.
Insect Biochem.Mol.Biol., 142, 2022
|
|
7NIH
 
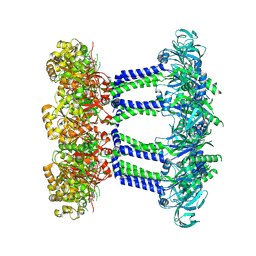 | | Wzc-K540M MgADP C8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
8HE5
 
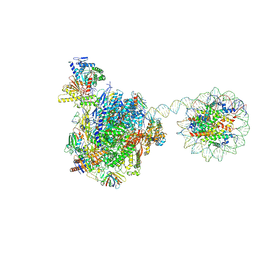 | | RNA polymerase II elongation complex bound with Rad26 and Elf1, stalled at SHL(-3.5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA repair protein, DNA-directed RNA polymerase subunit, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Kinoshita, C, Saotome, M, Kagawa, W, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.95 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7NII
 
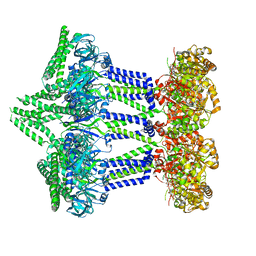 | | Wzc-K540M MgADP C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
8HL7
 
 | |
6X8H
 
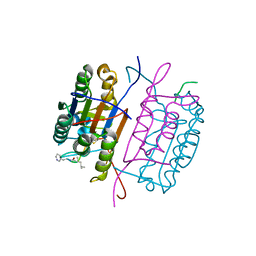 | | Caspase-8 in complex with AOMK inhibitor, Ac-DW3-KE, forms tetrahedral adduct | | Descriptor: | Ac-DW3-KE, Caspase-8 | | Authors: | Solania, A, Xu, J.H, Wolan, D.W. | | Deposit date: | 2020-06-01 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Caspase-8 in complex with AOMK inhibitor, Ac-DW3-KE, forms tetrahedral adduct
To Be Published
|
|
4KGX
 
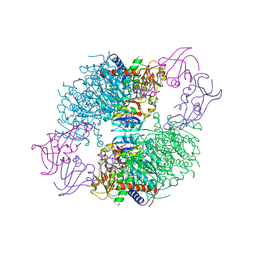 | | The R state structure of E. coli ATCase with CTP bound | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Cockrell, G.M, Zheng, Y, Guo, W, Peterson, A.W, Kantrowitz, E.R. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | New Paradigm for Allosteric Regulation of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 52, 2013
|
|
6X8L
 
 | |
6X8J
 
 | |
6X8I
 
 | |
8HNJ
 
 | | Domain-stabilized glutamine-binding protein | | Descriptor: | GLUTAMINE, Glutamine ABC transporter, periplasmic glutamine-binding protein GlnH | | Authors: | Choi, S.H, Park, J.H, Seo, M.H, Park, K.W, Lee, W.K. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Domain-wise dissection of thermal stability enhancement in multidomain proteins.
Int.J.Biol.Macromol., 237, 2023
|
|
