2UVO
 
 | | High Resolution Crystal Structure of Wheat Germ Agglutinin in Complex with N-Acetyl-D-Glucosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ ISOLECTIN 1, ... | | Authors: | Schwefel, D, Wittmann, V, Diederichs, K, Welte, W. | | Deposit date: | 2007-03-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
1SO2
 
 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 3B In COMPLEX WITH A DIHYDROPYRIDAZINE INHIBITOR | | Descriptor: | 1-DEOXY-1-[(2-HYDROXYETHYL)(NONANOYL)AMINO]HEXITOL, 6-(4-{[2-(3-IODOBENZYL)-3-OXOCYCLOHEX-1-EN-1-YL]AMINO}PHENYL)-5-METHYL-4,5-DIHYDROPYRIDAZIN-3(2H)-ONE, MAGNESIUM ION, ... | | Authors: | Scapin, G, Patel, S.B, Chung, C, Varnerin, J.P, Edmondson, S.D, Mastracchio, A, Parmee, E.R, Becker, J.W, Singh, S.B, Van Der Ploeg, L.H, Tota, M.R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Phosphodiesterase 3B: Atomic Basis for Substrate and Inhibitor Specificity
Biochemistry, 43, 2004
|
|
2UWH
 
 | | Cytochrome P450 BM3 mutant in complex with palmitic acid | | Descriptor: | BIFUNCTIONAL P-450: NADPH-P450 REDUCTASE, PALMITIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Huang, W.-C, Joyce, M.G, Westlake, A.C.G, Roberts, G.C.K, Moody, P.C.E. | | Deposit date: | 2007-03-21 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Filling a Hole in Cytochrome P450 Bm3 Improves Substrate Binding and Catalytic Efficiency.
J.Mol.Biol., 373, 2007
|
|
7AQQ
 
 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (membrane core) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-22 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7AQR
 
 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (peripheral arm) | | Descriptor: | Acyl carrier protein 2, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-22 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7ARC
 
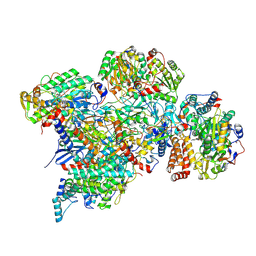 | | Cryo-EM structure of Polytomella Complex-I (peripheral arm) | | Descriptor: | 13 kDa, 18 kDa, 24 kDa, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7AR9
 
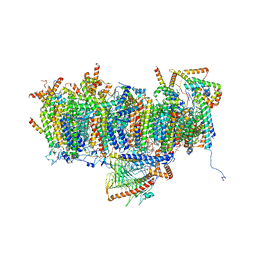 | | Cryo-EM structure of Polytomella Complex-I (membrane arm) | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 15 kDa, AGGG, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
3M32
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | Deposit date: | 2010-03-08 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
7ARD
 
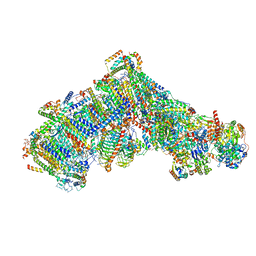 | | Cryo-EM structure of Polytomella Complex-I (complete composition) | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 13 kDa, 15 kDa, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
2UZP
 
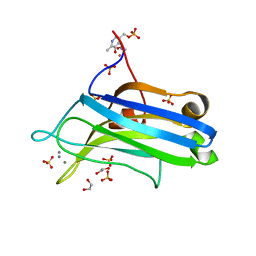 | | Crystal structure of the C2 domain of human protein kinase C gamma. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Pike, A.C.W, Amos, A, Johansson, C, Sobott, F, Savitsky, P, Berridge, G, Fedorov, O, Umeano, C, Gorrec, F, Bunkoczi, G, Debreczeni, J, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2007-04-30 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of C2 Domain of Protein Kinase C Gamma
To be Published
|
|
1SL6
 
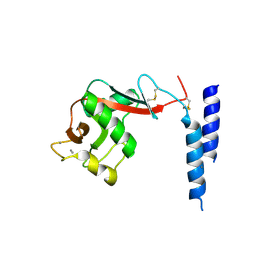 | | Crystal Structure of a fragment of DC-SIGNR (containg the carbohydrate recognition domain and two repeats of the neck) complexed with Lewis-x. | | Descriptor: | C-type lectin DC-SIGNR, CALCIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
5ZRZ
 
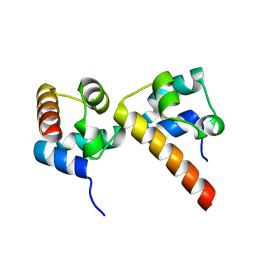 | | Crystal Structure of EphA5/SAMD5 Complex | | Descriptor: | Ephrin type-A receptor 5, Sterile alpha motif domain-containing protein 5 | | Authors: | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
7ZWO
 
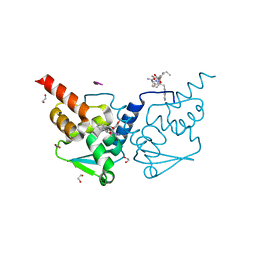 | | Crystal structure of human BCL6 BTB domain in complex with compound 2 | | Descriptor: | (5~{S},7~{R})-5-(4-chlorophenyl)-7-(2,3,4-trimethoxyphenyl)-4,5,6,7-tetrahydro-[1,2,4]triazolo[1,5-a]pyrimidine, 1,2-ETHANEDIOL, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWS
 
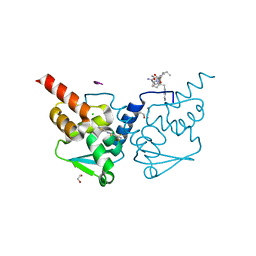 | | Crystal structure of human BCL6 BTB domain in complex with compound 13 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-cyano-6-thiophen-2-yl-4-(trifluoromethyl)pyridin-2-yl]sulfanyl-2-phenyl-ethanoic acid, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWR
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 11 | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-[[(6~{S})-3-cyano-6-methyl-4-(trifluoromethyl)-5,6,7,8-tetrahydroquinolin-2-yl]sulfanyl]ethanoylamino]ethanoic acid, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWX
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 19 | | Descriptor: | 1,2-ETHANEDIOL, 6-[1,3-benzodioxol-5-ylmethyl(methyl)amino]-1-~{tert}-butyl-5~{H}-pyrazolo[3,4-d]pyrimidin-4-one, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWZ
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 22 | | Descriptor: | 1,2-ETHANEDIOL, ALA-TRP-VAL-ILE-PRO-ALA, B-cell lymphoma 6 protein, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
6A0I
 
 | |
1IYL
 
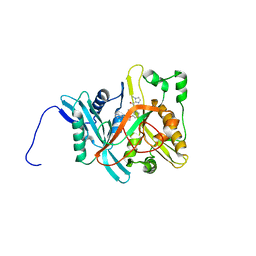 | | Crystal Structure of Candida albicans N-myristoyltransferase with Non-peptidic Inhibitor | | Descriptor: | (1-METHYL-1H-IMIDAZOL-2-YL)-(3-METHYL-4-{3-[(PYRIDIN-3-YLMETHYL)-AMINO]-PROPOXY}-BENZOFURAN-2-YL)-METHANONE, Myristoyl-CoA:Protein N-Myristoyltransferase | | Authors: | Sogabe, S, Fukami, T.A, Morikami, K, Shiratori, Y, Aoki, Y, D'Arcy, A, Winkler, F.K, Banner, D.W, Ohtsuka, T. | | Deposit date: | 2002-08-29 | | Release date: | 2002-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structures of Candida albicans N-Myristoyltransferase with Two Distinct Inhibitors
CHEM.BIOL., 9, 2002
|
|
7NRT
 
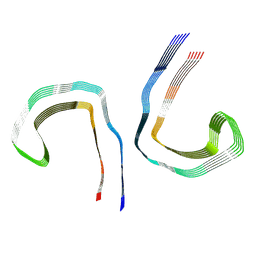 | | Conformation 2 of straight filament from primary age-related tauopathy brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7NRX
 
 | | Straight filament from Alzheimer's disease with PET ligand APN-1607 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7NRS
 
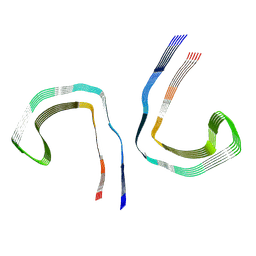 | | Conformation 1 of straight filament from primary age-related tauopathy brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7NRV
 
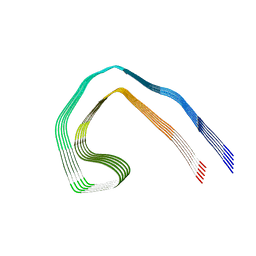 | | Paired helical filament from Alzheimer's disease with PET ligand APN-1607 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7NQ8
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH N-ethyl-3-(1-methyl-1H-1,2,3-triazol-4-yl)-4-(pyridin-2-ylmethoxy)benzamide | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, N-ethyl-3-(1-methyl-1H-1,2,3-triazol-4-yl)-4-(pyridin-2-ylmethoxy)benzamide | | Authors: | Chung, C.W. | | Deposit date: | 2021-03-01 | | Release date: | 2021-03-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Template-Hopping Approach Leads to Potent, Selective, and Highly Soluble Bromo and Extraterminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7NRQ
 
 | | Paired helical filament from primary age-related tauopathy brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
