7UG6
 
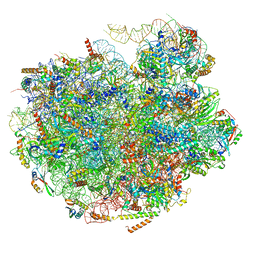 | | Cryo-EM structure of pre-60S ribosomal subunit, unmethylated G2922 | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Yelland, J.N, Bravo, J.P.K, Black, J.J.B, Taylor, D.W, Johnson, A.W. | | Deposit date: | 2022-03-24 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A single 2'-O-methylation of ribosomal RNA gates assembly of a functional ribosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5K2A
 
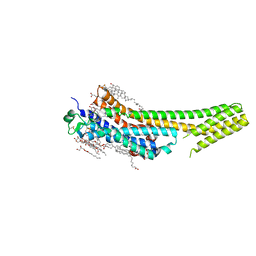 | | 2.5 angstrom A2a adenosine receptor structure with sulfur SAD phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
4YJY
 
 | |
1L02
 
 | |
1L09
 
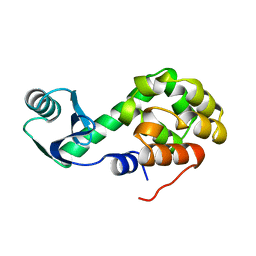 | |
4ZY6
 
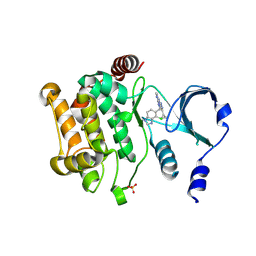 | |
5ZLU
 
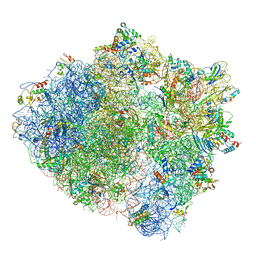 | | Ribosome Structure bound to ABC-F protein. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Su, W.X, Kumar, V, Ero, R, Andrew, S.W.W, Jian, S, Yong-Gui, G. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ribosome protection by antibiotic resistance ATP-binding cassette protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7UL1
 
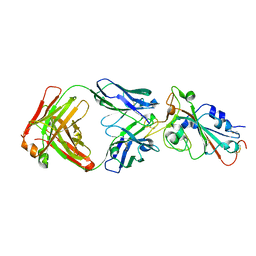 | | Crystal structure of SARS-CoV-2 RBD in complex with the neutralizing IGHV3-53-encoded antibody EH3 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH3, Light chain of EH3, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
7UL0
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with the ridge-binding nAb EH8 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH8, ... | | Authors: | Chen, Y, Tolbert, W.D, Bai, X, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
8SB6
 
 | | Structure of human BRD2-BD1 bound to a histone H4 acetyl-methyllysine peptide | | Descriptor: | Bromodomain containing 2, Histone H4 | | Authors: | Connor, L.J, Ekundayo, B.E, Lu-Culligan, W.J, Simon, M.D, Bleichert, F. | | Deposit date: | 2023-04-02 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acetyl-methyllysine marks chromatin at active transcription start sites.
Nature, 622, 2023
|
|
1L12
 
 | |
1L16
 
 | |
1L03
 
 | | CONTRIBUTIONS OF HYDROGEN BONDS OF THR 157 TO THE THERMODYNAMIC STABILITY OF PHAGE T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Dao-Pin, S, Wilson, K, Alber, T, Matthews, B.W. | | Deposit date: | 1988-02-05 | | Release date: | 1988-04-16 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme.
Nature, 330, 1987
|
|
1RTS
 
 | | THYMIDYLATE SYNTHASE FROM RAT IN TERNARY COMPLEX WITH DUMP AND TOMUDEX | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE, TOMUDEX | | Authors: | Sotelo-Mundo, R.R, Ciesla, J, Dzik, J.M, Rode, W, Maley, F, Maley, G, Hardy, L.W, Montfort, W.R. | | Deposit date: | 1998-06-19 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of rat thymidylate synthase inhibited by Tomudex, a potent anticancer drug.
Biochemistry, 38, 1999
|
|
8SCL
 
 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 6h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCN
 
 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 24h (Product State) | | Descriptor: | CALCIUM ION, DIPHOSPHATE, DNA polymerase I, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCQ
 
 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 2h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCP
 
 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 1h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCS
 
 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 8h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCR
 
 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 4h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCT
 
 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 24h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCU
 
 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 48h (Product State) | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
2CEH
 
 | | Phosphorylation of the Cytoplasmic Tail of Tissue Factor and its Role in Modulating Structure and Binding Affinity | | Descriptor: | TISSUE FACTOR | | Authors: | Sen, M, Agrawal, S, Craft, J.W, Ruf, W, Legge, G.B. | | Deposit date: | 2006-02-06 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic Characterization of Successive Phosphorylation of the Tissue Factor Cytoplasmic Region.
Open Spectrosc.J., 3, 2009
|
|
1L01
 
 | |
1L07
 
 | |
