1H0M
 
 | | Three-dimensional structure of the quorum sensing protein TraR bound to its autoinducer and to its target DNA | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, 5'-D(*AP*TP*GP*TP*GP*CP*AP*GP*AP*TP *CP*TP*GP*CP*AP*CP*AP*T)-3', Transcriptional activator protein TraR | | Authors: | Vannini, A, Volpari, C, Gargioli, C, Muraglia, E, Cortese, R, De Francesco, R, Neddermann, P, Di Marco, S. | | Deposit date: | 2002-06-25 | | Release date: | 2002-08-29 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of the Quorum Sensing Protein Trar Bound to its Autoinducer and Target DNA
Embo J., 21, 2002
|
|
1W3C
 
 | | Crystal structure of the Hepatitis C Virus NS3 Protease in complex with a peptidomimetic inhibitor | | Descriptor: | 3-({(2S)-2-[({(1R)-1-[({(1R)-1-[(R)-CARBOXY(HYDROXY)METHYL]-3,3-DIFLUOROPROPYL}AMINO)CARBONYL]-3-METHYLBUTYL}AMINO)CARB ONYL]-2,3-DIHYDRO-1H-INDOL-2-YL}METHYL)THIOPHENE-2-CARBOXYLIC ACID, 3-({(2S)-2-[({(1S)-1-[({(1S)-1-[(R)-CARBOXY(HYDROXY)METHYL]-3,3-DIFLUOROPROPYL}AMINO)CARBONYL]-3-METHYLBUTYL}AMINO)CARBONYL]-2,3-DIHYDRO-1H-INDOL-2-YL}METHYL)THIOPHENE-2-CARBOXYLIC ACID, NONSTRUCTURAL PROTEIN NS4A (P4), ... | | Authors: | Di Marco, S, Volpari, C. | | Deposit date: | 2004-07-14 | | Release date: | 2004-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Design and Enzyme-Bound Crystal Structure of Indoline Based Peptidomimetic Inhibitors of Hepatitis C Virus Ns3 Protease
J.Med.Chem., 47, 2004
|
|
2V5X
 
 | | Crystal structure of HDAC8-inhibitor complex | | Descriptor: | (2R)-N~8~-HYDROXY-2-{[(5-METHOXY-2-METHYL-1H-INDOL-3-YL)ACETYL]AMINO}-N~1~-[2-(2-PHENYL-1H-INDOL-3-YL)ETHYL]OCTANEDIAMIDE, HISTONE DEACETYLASE 8, POTASSIUM ION, ... | | Authors: | Di Marco, S, Vannini, A, Volpari, C, Gallinari, P, Jones, P, Mattu, M, Carfi, A, Defrancesco, R, Steinkuhler, C. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate Binding to Histone Deacetylases as Revealed by Crystal Structure of Hdac8-Substrate Complex
Embo Rep., 8, 2007
|
|
2V5W
 
 | | Crystal structure of HDAC8-substrate complex | | Descriptor: | GLYCYL-GLYCYL-GLYCINE, HISTONE DEACETYLASE 8, PEPTIDIC SUBSTRATE, ... | | Authors: | Di Marco, S, Vannini, A, Volpari, C. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Binding to Histone Deacetylases as Revealed by Crystal Structure of Hdac8-Substrate Complex
Embo Rep., 8, 2007
|
|
1W22
 
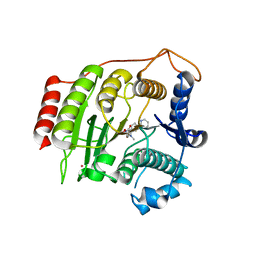 | | Crystal structure of inhibited human HDAC8 | | Descriptor: | HISTONE DEACETYLASE 8, N-HYDROXY-4-(METHYL{[5-(2-PYRIDINYL)-2-THIENYL]SULFONYL}AMINO)BENZAMIDE, POTASSIUM ION, ... | | Authors: | Vannini, A, Volpari, C, Caroli Casavola, E, Di Marco, S. | | Deposit date: | 2004-06-25 | | Release date: | 2004-09-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Eukaryotic Zn-Dependent Histone Deacetylase,Human Hdac8,Complexed with a Hydroxamic Acid Inhibitor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2XTJ
 
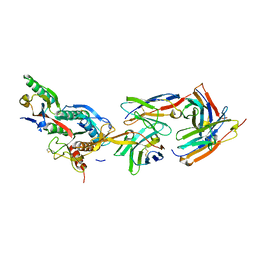 | | The crystal structure of PCSK9 in complex with 1D05 Fab | | Descriptor: | CALCIUM ION, FAB FROM A HUMAN MONOCLONAL ANTIBODY, 1D05, ... | | Authors: | Di Marco, S, Volpari, C, Carfi, A. | | Deposit date: | 2010-10-10 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Pcsk9-Binding Antibody that Structurally Mimics the Egf(A) Domain of Ldl-Receptor Reduces Ldl Cholesterol in Vivo.
J.Lipid Res., 52, 2011
|
|
1DY9
 
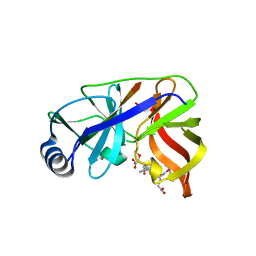 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor I) | | Descriptor: | N-(tert-butoxycarbonyl)-L-alpha-glutamyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1DY8
 
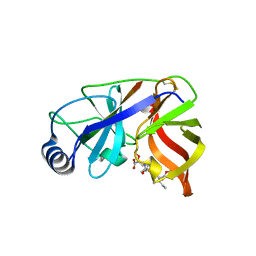 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor II) | | Descriptor: | N-[(benzyloxy)carbonyl]-L-isoleucyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70) | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1DXP
 
 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (apo structure) | | Descriptor: | GLYCEROL, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-13 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
2BRK
 
 | |
2BRL
 
 | |
