2KQN
 
 | |
2KOL
 
 | |
2GOW
 
 | | Solution structure of BC059385 from Homo sapiens | | Descriptor: | Ubiquitin-like protein 3 | | Authors: | Volkman, B.F, de la Cruz, N.B, Lytle, B.L, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-14 | | Release date: | 2006-04-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a membrane-anchored ubiquitin-fold (MUB) protein from Homo sapiens.
Protein Sci., 16, 2007
|
|
2I9Y
 
 | | Solution structure of Arabidopsis thaliana protein At1g70830, a member of the major latex protein family | | Descriptor: | major latex protein-like protein 28 or MLP-like protein 28 | | Authors: | Volkman, B.F, de la Cruz, N.B, Lytle, B.L, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-06 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structures of two Arabidopsis thaliana major latex proteins represent novel helix-grip folds.
Proteins, 76, 2009
|
|
2L47
 
 | |
2LC7
 
 | |
2L48
 
 | |
2JP1
 
 | |
2JQV
 
 | |
2K04
 
 | |
2K03
 
 | |
2LGR
 
 | |
2LC6
 
 | |
2KQR
 
 | |
5I7Z
 
 | | Crystal structure of a Par-6 PDZ-Crumbs 3 C-terminal peptide complex | | Descriptor: | Crb-3, DI(HYDROXYETHYL)ETHER, LD29223p | | Authors: | Whitney, D.S, Peterson, F.C, Prehoda, K.E, Volkman, B.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Binding of Crumbs to the Par-6 CRIB-PDZ Module Is Regulated by Cdc42.
Biochemistry, 55, 2016
|
|
6CWS
 
 | |
4WVO
 
 | | An engineered PYR1 mandipropamid receptor in complex with mandipropamid and HAB1 | | Descriptor: | (2S)-2-(4-chlorophenyl)-N-{2-[3-methoxy-4-(prop-2-yn-1-yloxy)phenyl]ethyl}-2-(prop-2-yn-1-yloxy)ethanamide, Abscisic acid receptor PYR1, MAGNESIUM ION, ... | | Authors: | Peterson, F.C, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-02-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Agrochemical control of plant water use using engineered abscisic acid receptors.
Nature, 520, 2015
|
|
5KTF
 
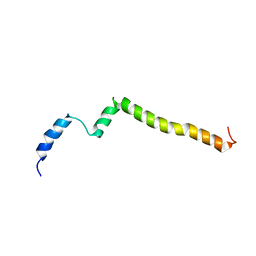 | | Structure of the C-terminal transmembrane domain of scavenger receptor BI (SR-BI) | | Descriptor: | Scavenger receptor class B member 1 | | Authors: | Chadwick, A.C, Peterson, F.C, Volkman, B.F, Sahoo, D. | | Deposit date: | 2016-07-11 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the C-Terminal Transmembrane Domain of the HDL Receptor, SR-BI, and a Functionally Relevant Leucine Zipper Motif.
Structure, 25, 2017
|
|
4A1M
 
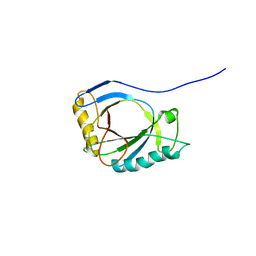 | | NMR Structure of protoporphyrin-IX bound murine p22HBP | | Descriptor: | HEME-BINDING PROTEIN 1 | | Authors: | Goodfellow, B.J, Dias, J.S, Macedo, A.L, Ferreira, G.C, Peterson, F.C, Volkman, B.F, Duarte, I.C.N. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of Protoporphyrin-Ix Bound Murine P22Hbp
To be Published
|
|
4F4J
 
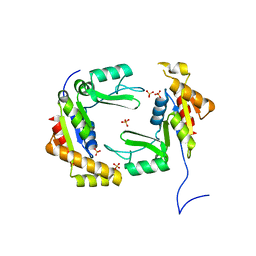 | | Conversion of the enzyme guanylate kinase into a mitotic spindle orienting protein by a single mutation that inhibits gmp- induced closing | | Descriptor: | Guanylate kinase, SULFATE ION | | Authors: | Johnston, C.A, Whitney, D, Volkman, B.F, Prehoda, K.E. | | Deposit date: | 2012-05-10 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conversion of the enzyme guanylate kinase into a mitotic-spindle orienting protein by a single mutation that inhibits GMP-induced closing.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8UEK
 
 | |
8EY0
 
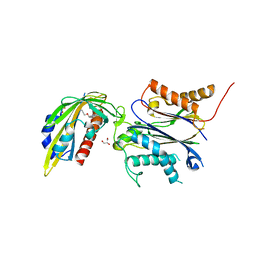 | | Structure of an orthogonal PYR1*:HAB1* chemical-induced dimerization module in complex with mandipropamid | | Descriptor: | (2S)-2-(4-chlorophenyl)-N-{2-[3-methoxy-4-(prop-2-yn-1-yloxy)phenyl]ethyl}-2-(prop-2-yn-1-yloxy)ethanamide, Abscisic acid receptor PYR1, GLYCEROL, ... | | Authors: | Park, S.-Y, Volkman, B.F, Cutler, S.R, Peterson, F.C. | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An orthogonalized PYR1-based CID module with reprogrammable ligand-binding specificity.
Nat.Chem.Biol., 20, 2024
|
|
3NJ1
 
 | | X-ray crystal structure of the PYL2(V114I)-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, GLYCEROL, ... | | Authors: | Peterson, F.C, Burgie, E.S, Bingman, C.A, Volkman, B.F, Phillips Jr, G.N, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NJ0
 
 | | X-ray crystal structure of the PYL2-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peterson, F.C, Burgie, E.S, Bingman, C.A, Volkman, B.F, Phillips Jr, G.N, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6NWB
 
 | | PYL10 bound to the selective agonist hexabactin | | Descriptor: | Abscisic acid receptor PYL10, N-{[(4-cyanophenyl)methyl]sulfonyl}-1-(thiophen-3-yl)cyclohexane-1-carboxamide | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2019-02-06 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | PYL10 bound to the selective agonist hexabactin
To Be Published
|
|
