4UQ8
 
 | | Electron cryo-microscopy of bovine Complex I | | Descriptor: | ACYL CARRIER PROTEIN, MITOCHONDRIAL, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2014-06-21 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.95 Å) | | Cite: | Architecture of Mammalian Respiratory Complex I.
Nature, 515, 2014
|
|
4CKD
 
 | |
2XOW
 
 | | Structure of GlpG in complex with a mechanism-based isocoumarin inhibitor | | Descriptor: | 5-AMINO-2-(2-METHOXY-2-OXOETHYL)BENZOIC ACID, RHOMBOID PROTEASE GLPG, nonyl beta-D-glucopyranoside | | Authors: | Vinothkumar, K.R, Strisovsky, K, Andreeva, A, Christova, Y, Verhelst, S, Freeman, M. | | Deposit date: | 2010-08-24 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Structural Basis for Catalysis and Substrate Specificity of a Rhomboid Protease
Embo J., 29, 2010
|
|
2XTU
 
 | |
2XOV
 
 | | Crystal Structure of E.coli rhomboid protease GlpG, native enzyme | | Descriptor: | GLYCEROL, RHOMBOID PROTEASE GLPG, nonyl beta-D-glucopyranoside | | Authors: | Vinothkumar, K.R, Strisovsky, K, Andreeva, A, Christova, Y, Verhelst, S, Freeman, M. | | Deposit date: | 2010-08-24 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structural basis for catalysis and substrate specificity of a rhomboid protease.
EMBO J., 29, 2010
|
|
2XTV
 
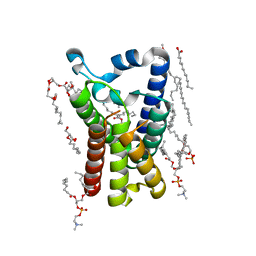 | |
7W8J
 
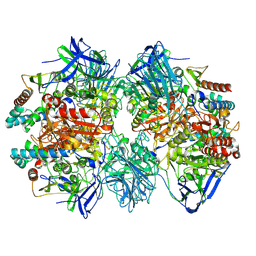 | | Dimethylformamidase, 2x(A2B2) | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Vinothkumar, K.R, Subramanian, R, Arya, C, Ramanathan, G. | | Deposit date: | 2021-12-07 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dimethylformamidase with a Unique Iron Center
To Be Published
|
|
3ZOT
 
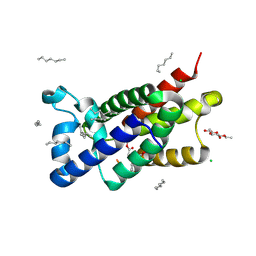 | | Structure of E.coli rhomboid protease GlpG in complex with monobactam L29 (data set 2) | | Descriptor: | CHLORIDE ION, RHOMBOID PROTEASE GLPG, nonyl beta-D-glucopyranoside, ... | | Authors: | Vinothkumar, K.R, Pierrat, O.A, Large, J.M, Freeman, M. | | Deposit date: | 2013-02-24 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structure of Rhomboid Protease in Complex with Beta-Lactam Inhibitors Defines the S2' Cavity.
Structure, 21, 2013
|
|
3ZMH
 
 | | Structure of E.coli rhomboid protease GlpG in complex with monobactam L62 | | Descriptor: | CHLORIDE ION, CYCLOPENTYL 2-OXO-4-PHENYLAZETIDINE-1-CARBOXYLATE, RHOMBOID PROTEASE GLPG, ... | | Authors: | Vinothkumar, K.R, Pierrat, O.A, Large, J.M, Freeman, M. | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of rhomboid protease in complex with beta-lactam inhibitors defines the S2' cavity.
Structure, 21, 2013
|
|
3ZMJ
 
 | | Structure of E.coli rhomboid protease GlpG in complex with monobactam L61 | | Descriptor: | 2-methylpropyl N-[(1R)-3-oxidanylidene-1-phenyl-propyl]carbamate, CHLORIDE ION, RHOMBOID PROTEASE GLPG, ... | | Authors: | Vinothkumar, K.R, Pierrat, O, Large, J.M, Freeman, M. | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Rhomboid Protease in Complex with Beta-Lactam Inhibitors Defines the S2' Cavity.
Structure, 21, 2013
|
|
3ZEB
 
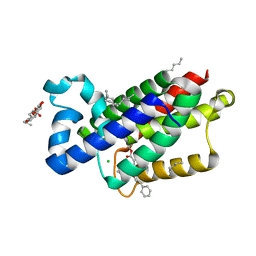 | | A complex of GlpG with isocoumarin inhibitor covalently bonded to serine 201 and histidine 150 | | Descriptor: | 2-phenylethyl 2-(4-azanyl-2-methanoyl-phenyl)ethanoate, CHLORIDE ION, RHOMBOID PROTEASE GLPG, ... | | Authors: | Vinothkumar, K.R, voskya, O, Kuettler, E.V, Brouwer, A.J, Liskamp, R.M.J, Verhelst, S.H.L. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activity-Based Probes for Rhomboid Proteases Discovered in a Mass Spectrometry-Based Assay.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZMI
 
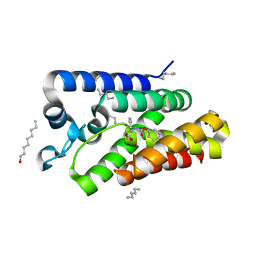 | | Structure of E.coli rhomboid protease GlpG in complex with monobactam L29 | | Descriptor: | RHOMBOID PROTEASE GLPG, nonyl beta-D-glucopyranoside, phenyl N-[(1R)-3-oxidanylidene-1-phenyl-propyl]carbamate | | Authors: | Vinothkumar, K.R, Pierrat, O.A, Large, J.M, Freeman, M. | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Rhomboid Protease in Complex with Beta-Lactam Inhibitors Defines the S2' Cavity.
Structure, 21, 2013
|
|
5LDX
 
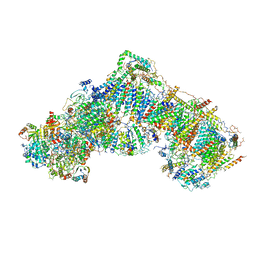 | | Structure of mammalian respiratory Complex I, class3. | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2016-06-28 | | Release date: | 2016-09-14 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure of mammalian respiratory complex I.
Nature, 536, 2016
|
|
5LC5
 
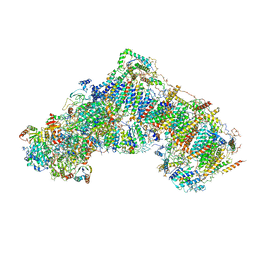 | | Structure of mammalian respiratory Complex I, class2 | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2016-06-19 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Structure of mammalian respiratory complex I.
Nature, 536, 2016
|
|
5LQY
 
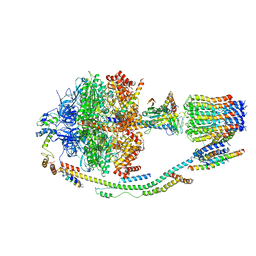 | | Structure of F-ATPase from Pichia angusta, in state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase OSCP subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5LQX
 
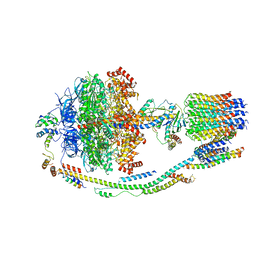 | | Structure of F-ATPase from Pichia angusta, state3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase AAP1 subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5LDW
 
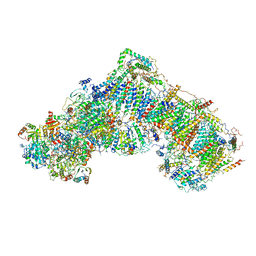 | | Structure of mammalian respiratory Complex I, class1 | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2016-06-28 | | Release date: | 2016-09-07 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structure of mammalian respiratory complex I.
Nature, 536, 2016
|
|
5LQZ
 
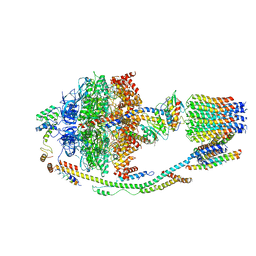 | | Structure of F-ATPase from Pichia angusta, state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase OSCP subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5MT6
 
 | |
5MT8
 
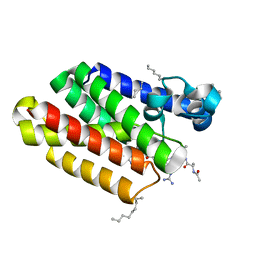 | |
5MT7
 
 | |
7FD9
 
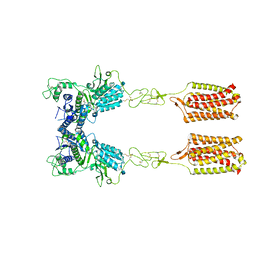 | | Thermostabilised full length human mGluR5-5M with orthosteric antagonist, LY341495 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5 | | Authors: | Vinothkumar, K.R, Cannone, G, Lebon, G. | | Deposit date: | 2021-07-16 | | Release date: | 2021-09-08 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Agonists and allosteric modulators promote signaling from different metabotropic glutamate receptor 5 conformations.
Cell Rep, 36, 2021
|
|
7FD8
 
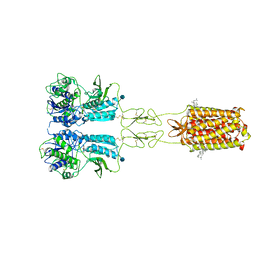 | | Thermostabilised full length human mGluR5-5M bound with L-quisqualic acid | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Vinothkumar, K.R, Cannone, G, Lebon, G. | | Deposit date: | 2021-07-16 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Agonists and allosteric modulators promote signaling from different metabotropic glutamate receptor 5 conformations.
Cell Rep, 36, 2021
|
|
7Y0D
 
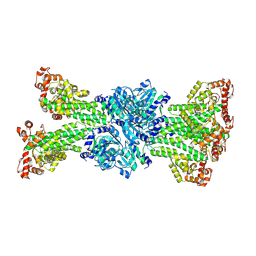 | |
3FI1
 
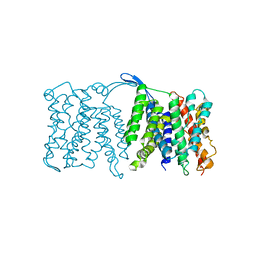 | | NhaA dimer model | | Descriptor: | Na(+)/H(+) antiporter nhaA | | Authors: | Appel, M, Hizlan, D, Vinothkumar, K.R, Ziegler, C, Kuehlbrandt, W. | | Deposit date: | 2008-12-10 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Conformations of NhaA, the Na/H exchanger from Escherichia coli, in the pH-activated and ion-translocating states
J.Mol.Biol., 386, 2009
|
|
