4AIZ
 
 | | Crystallographic structure of 3mJL2 from the germinal line lambda 3 | | Descriptor: | 1,5:6,10-dianhydro-3,4,7,8-tetradeoxy-2,9-bis-C-(hydroxymethyl)-L-manno-decitol, CITRIC ACID, SULFATE ION, ... | | Authors: | Villalba, M.I, Luna, O.D, Rudino-Pinera, E, Sanchez, R, Sanchez-Lopez, R, Rojas-Trejo, S, Olamendi-Portugal, T, Fernandez-Velasco, D.A, Becerril, B. | | Deposit date: | 2012-02-15 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Site-Directed Mutagenesis Reveals Regions Implicated in the Stability and Fiber Formation of Human Lambda3R Light Chains.
J.Biol.Chem., 290, 2015
|
|
4AJ0
 
 | | Crystallographic structure of an amyloidogenic variant, 3rCW, of the germinal line lambda 3 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, ACETATE ION, GERMINAL LINE LAMBDA 3 3RCW VARIANT | | Authors: | Villalba, M.I, Luna, O.D, Rudino-Pinera, E, Sanchez, R, Sanchez-Lopez, R, Rojas-Trejo, S, Olamendi-Portugal, T, Fernandez-Velasco, D.A, Becerril, B. | | Deposit date: | 2012-02-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Site-Directed Mutagenesis Reveals Regions Implicated in the Stability and Fiber Formation of Human Lambda3R Light Chains.
J.Biol.Chem., 290, 2015
|
|
4AIX
 
 | | Crystallographic structure of an amyloidogenic variant, 3rC34Y, of the germinal line lambda 3 | | Descriptor: | IG LAMBDA CHAIN V-IV REGION BAU | | Authors: | Villalba, M.I, Luna, O.D, Rudino-Pinera, E, Sanchez, R, Sanchez-Lopez, R, Rojas-Trejo, S, Olamendi-Portugal, T, Fernandez-Velasco, D.A, Becerril, B. | | Deposit date: | 2012-02-15 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Site-Directed Mutagenesis Reveals Regions Implicated in the Stability and Fiber Formation of Human Lambda3R Light Chains.
J.Biol.Chem., 290, 2015
|
|
4UV9
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Ethyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(4aS,10aS)-4a-[(1S,3E)-3-imino-1-phenylpentyl]-7,8-dimethyl-2,4-dioxo-1,3,4,4a,5,10a-hexahydrobenzo[g]pteridin-10(2H)-yl]pentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
4UVB
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Methyl-Tranylcypromine (1S,2R) | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,7aS)-1-amino-1,10,11-trimethyl-4,6-dioxo-3-phenyl-2,3,5,6,7,7a-hexahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
4UV8
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Benzyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [[(2R,3S,4S)-5-[(4aS,10aS)-4a-[(1S)-3-azanylidene-1,4-diphenyl-butyl]-7,8-dimethyl-2,4-bis(oxidanylidene)-5,10a-dihydro-1H-benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentoxy]-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
2C4G
 
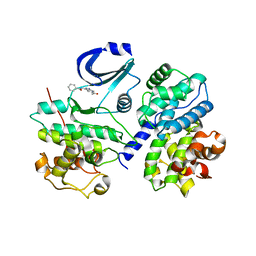 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-533514 | | Descriptor: | (3Z)-5-ACETYL-3-(BENZOYLIMINO)-3,6-DIHYDROPYRROLO[3,4-C]PYRAZOL-5-IUM, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Cameron, A, Fogliatto, G, Pevarello, P, Fancelli, D, Vulpetti, A, Amici, R, Villa, M, Pittala, V, Ciomei, M, Mercurio, C, Bischoff, J.R, Roletto, F, Varasi, M, Brasca, M.G. | | Deposit date: | 2005-10-19 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 3-Amino-1,4,5,6-Tetrahydropyrrolo[3,4-C]Pyrazoles: A New Class of Cdk2 Inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4UVC
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Phenyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4S)-5-[5-[(1S)-1-azanyl-1,3-diphenyl-propyl]-7,8-dimethyl-2,4-bis(oxidanylidene)-4aH-benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentyl] hydrogen phosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
4UVA
 
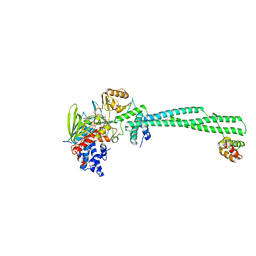 | | LSD1(KDM1A)-CoREST in complex with 1-Methyl-Tranylcypromine (1R,2S) | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(4aS)-4a-[(1S,3E)-3-imino-1-phenylbutyl]-7,8-dimethyl-2,4-dioxo-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]pentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
1VYZ
 
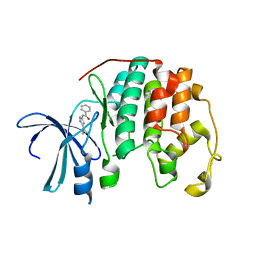 | | Structure of CDK2 complexed with PNU-181227 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-(5-CYCLOPROPYL-1H-PYRAZOL-3-YL)BENZAMIDE | | Authors: | Pevarello, P, Brasca, M.G, Amici, R, Orsini, P, Traquandi, G, Corti, L, Piutti, C, Sansonna, P, Villa, M, Pierce, B.S, Pulici, M, Giordano, G, Martina, K, Lfritzen, E, Nugent, R.A, Casale, E, Cameron, A, Ciomei, M, Roletto, F, Isacchi, A, Fogliatto, G, Pesenti, E, Pastori, W, Marsiglio, W, Leach, K.L, Clare, P.M, Fiorentini, F, Varasi, M, Vulpetti, A, Warpehoski, M.A. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 3-Aminopyrazole Inhibitors of Cdk2/Cyclin a as Antitumor Agents. 1. Lead Finding
J.Med.Chem., 47, 2004
|
|
1VYW
 
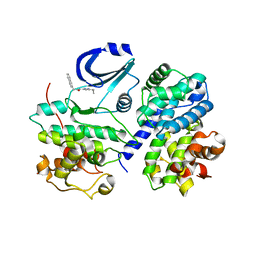 | | Structure of CDK2/Cyclin A with PNU-292137 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, N-(3-CYCLOPROPYL-1H-PYRAZOL-5-YL)-2-(2-NAPHTHYL)ACETAMIDE, ... | | Authors: | Pevarello, P, Brasca, M.G, Amici, R, Orsini, P, Traquandi, G, Corti, L, Piutti, C, Sansonna, P, Villa, M, Pierce, B.S, Pulici, M, Giordano, P, Martina, K, Fritzen, E.L, Nugent, R.A, Casale, E, Cameron, A, Ciomei, M, Roletto, F, Isacchi, A, Fogliatto, G, Pesenti, E, Pastori, W, Marsiglio, A, Leach, K.L, Clare, P.M, Fiorentini, F, Varasi, M, Vulpetti, A, Warpehoski, M.A. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Aminopyrazole Inhibitors of Cdk2/Cyclin a as Antitumor Agents. Part 1. Lead Finding
J.Med.Chem., 47, 2004
|
|
2BTR
 
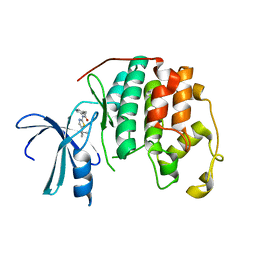 | | STRUCTURE OF CDK2 COMPLEXED WITH PNU-198873 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-(5-ISOPROPYL-THIAZOL-2-YL)-2-PYRIDIN-3-YL-ACETAMIDE | | Authors: | Vulpetti, A, Casale, E, Roletto, F, Amici, R, Villa, M, Pevarello, P. | | Deposit date: | 2005-06-06 | | Release date: | 2005-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Drug Design to the Discovery of New 2-Aminothiazole Cdk2 Inhibitors.
J.Mol.Graph.Model., 24, 2006
|
|
2BTS
 
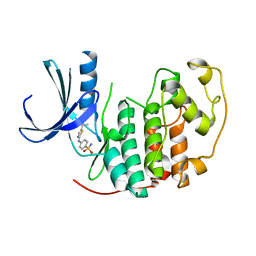 | | STRUCTURE OF CDK2 COMPLEXED WITH PNU-230032 | | Descriptor: | 4-[(5-ISOPROPYL-1,3-THIAZOL-2-YL)AMINO]BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Vulpetti, A, Casale, E, Roletto, F, Amici, R, Villa, M, Pevarello, P. | | Deposit date: | 2005-06-06 | | Release date: | 2005-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Drug Design to the Discovery of New 2-Aminothiazole Cdk2 Inhibitors.
J.Mol.Graph.Model., 24, 2006
|
|
9EP9
 
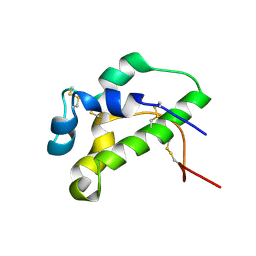 | | NMR solution structure of lipid transfer protei Sola l7 from tomato seeds | | Descriptor: | Non-specific lipid-transfer protein | | Authors: | Parron-Ballesteros, J, Mantin-Pedraz, L, G.Gordo, R, Mayorga, C, Villaba, M, Batanero, E, Pantoja-Uceda, D, Turnay, J. | | Deposit date: | 2024-03-18 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Long-chain fatty acids block allergic reaction against lipid transfer protein Sola l 7 from tomato seeds.
Protein Sci., 33, 2024
|
|
7OQH
 
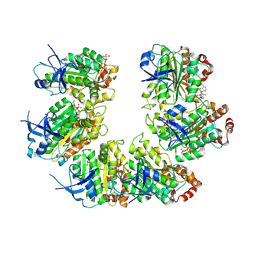 | | CryoEM structure of the transcription termination factor Rho from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Transcription termination factor Rho | | Authors: | Saridakis, E, Vishwakarma, R, Lai Kee Him, J, Martin, K, Simon, I, Cohen-Gonsaud, M, Coste, F, Bron, P, Margeat, E, Boudvillain, M. | | Deposit date: | 2021-06-03 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structure of transcription termination factor Rho from Mycobacterium tuberculosis reveals bicyclomycin resistance mechanism.
Commun Biol, 5, 2022
|
|
1SS3
 
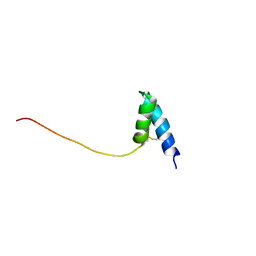 | | Solution structure of Ole e 6, an allergen from olive tree pollen | | Descriptor: | Pollen allergen Ole e 6 | | Authors: | Trevino, M.A, Garcia-Mayoral, M.F, Barral, P, Villalba, M, Santoro, J, Rico, M, Rodriguez, R, Bruix, M. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Ole e 6, a Major Allergen from Olive Tree Pollen.
J.Biol.Chem., 279, 2004
|
|
2XE0
 
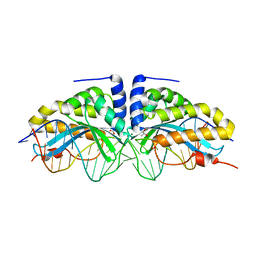 | | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus | | Descriptor: | 24MER DNA, ACETATE ION, I-CREI V2V3 VARIANT, ... | | Authors: | Munoz, I.G, Prieto, J, Subramanian, S, Coloma, J, Redondo, P, Villate, M, Merino, N, Marenchino, M, D'Abramo, M, Gervasio, F.L, Grizot, S, Daboussi, F, Smith, J, Chion-Sotine, I, Paques, F, Duchateau, P, Alibes, A, Stricher, F, Serrano, L, Blanco, F.J, Montoya, G. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Molecular Basis of Engineered Meganuclease Targeting of the Endogenous Human Rag1 Locus
Nucleic Acids Res., 39, 2011
|
|
1SM7
 
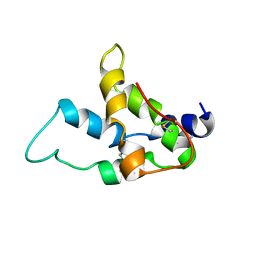 | | Solution structure of the recombinant pronapin precursor, BnIb. | | Descriptor: | recombinant Ib pronapin | | Authors: | Pantoja-Uceda, D, Palomares, O, Bruix, M, Villalba, M, Rodriguez, R, Rico, M, Santoro, J. | | Deposit date: | 2004-03-08 | | Release date: | 2005-02-01 | | Last modified: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: relationship to its allergenic properties.
Biochemistry, 43, 2004
|
|
2JON
 
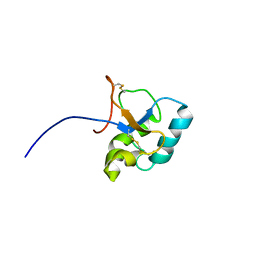 | | Solution structure of the C-terminal domain Ole e 9 | | Descriptor: | Beta-1,3-glucanase | | Authors: | Trevino, M.A, Palomares, O, Castrillo, I, Villalba, M, Rodriguez, R, Rico, M, Santoro, J, Bruix, M. | | Deposit date: | 2007-03-14 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of Ole e 9, a major allergen of olive pollen
Protein Sci., 17, 2008
|
|
2XAO
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with IP5 | | Descriptor: | INOSITOL-PENTAKISPHOSPHATE 2-KINASE, MYO-INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, ZINC ION | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5A74
 
 | | Crystal structure of the homing endonuclease I-CvuI in complex with its target (Sro1.3) in the presence of 2 mM Mn | | Descriptor: | 10MER DNA, 5'-D(*GP*AP*CP*GP*TP*TP*CP*TP*GP*AP)-3', 14MER DNA, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
5A0M
 
 | | THE CRYSTAL STRUCTURE OF I-SCEI IN COMPLEX WITH ITS TARGET DNA IN THE PRESENCE OF MN | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*TP*AP*GP*GP*GP*AP*TP*AP*AP)-3', 5'-D(*CP*AP*GP*GP*GP*TP*AP*AP*TP*AP*CP)-3', 5'-D(*CP*CP*CP*TP*AP*GP*CP*GP*TP)-3', ... | | Authors: | Prieto, J, Redondo, P, Merino, N, Villate, M, Blanco, F.J, Molina, R. | | Deposit date: | 2015-04-21 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the I-Scei Nuclease Complexed with its DsDNA Target and Three Catalytic Metal Ions.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6YOA
 
 | | Lig v 1 structure and the inflammatory response to the Ole e 1 protein family | | Descriptor: | Major pollen allergen Lig v 1, NICKEL (II) ION | | Authors: | Robledo-Retana, T, Bradley-Clark, J, Croll, T, Rose, R, Stagg, A, Villalba, M, Pickersgill, R. | | Deposit date: | 2020-04-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Lig v 1 structure and the inflammatory response to the Ole e 1 protein family.
Allergy, 75, 2020
|
|
5A77
 
 | | Crystal structure of the homing endonuclease I-CvuI in complex with I- CreI target (C1221) in the presence of 2 mM Mg revealing DNA cleaved | | Descriptor: | 10MER DNA, 5'-D(*GP*AP*CP*GP*TP*TP*TP*TP* GP*AP*DGP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', 14MER DNA, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
5A72
 
 | | Crystal structure of the homing endonuclease I-CvuI in complex with its target (Sro1.3) in the presence of 2 mM Ca | | Descriptor: | 24MER DNA, 5'-D(*DTP*CP*AP*GP*AP*AP*CP*GP*TP*CP*GP*TP*AP *DCP*GP*AP*CP*GP*TP*TP*CP*TP*GP*A)-3', CALCIUM ION, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
