5WHA
 
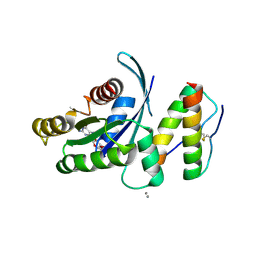 | | KRas G12V, bound to GDP and miniprotein 225-11 | | Descriptor: | CALCIUM ION, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Shim, S.Y, McGee, J.H, Lee, S.-J, Verdine, G.L. | | Deposit date: | 2017-07-16 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Exceptionally high-affinity Ras binders that remodel its effector domain.
J. Biol. Chem., 293, 2018
|
|
5WHB
 
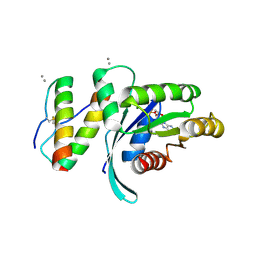 | | KRas G12V, bound to GDP and miniprotein 225-11(A30R) | | Descriptor: | CALCIUM ION, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Shim, S.Y, McGee, J.H, Lee, S.-J, Verdine, G.L. | | Deposit date: | 2017-07-16 | | Release date: | 2018-01-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Exceptionally high-affinity Ras binders that remodel its effector domain.
J. Biol. Chem., 293, 2018
|
|
5WLB
 
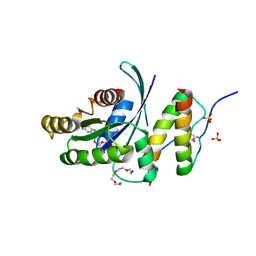 | | KRas G12V, bound to GppNHp and miniprotein 225-15a/b | | Descriptor: | 225-15 a, 225-15 b, GTPase KRas, ... | | Authors: | Shim, S.Y, McGee, J.H, Lee, S.-J, Verdine, G.L. | | Deposit date: | 2017-07-26 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Exceptionally high-affinity Ras binders that remodel its effector domain.
J. Biol. Chem., 293, 2018
|
|
5WHE
 
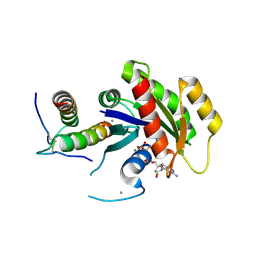 | | KRas G12V/D38P, bound to GppNHp and miniprotein 225-11 | | Descriptor: | CALCIUM ION, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Shim, S.Y, McGee, J.H, Lee, S.-J, Verdine, G.L. | | Deposit date: | 2017-07-16 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Exceptionally high-affinity Ras binders that remodel its effector domain.
J. Biol. Chem., 293, 2018
|
|
5WPL
 
 | | KRas G12V, bound to GppNHp and miniprotein 225-11 | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Lee, S.-J, Shim, S.Y, McGee, J.H, Verdine, G.L. | | Deposit date: | 2017-08-05 | | Release date: | 2018-01-03 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Exceptionally high-affinity Ras binders that remodel its effector domain.
J. Biol. Chem., 293, 2018
|
|
2F5P
 
 | | MutM crosslinked to undamaged DNA sampling A:T base pair IC2 | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*TP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*G*TP*CP*CP*AP*AP*GP*TP*CP*TP*AP*CP*C)-3', ZINC ION, ... | | Authors: | Banerjee, A, Santos, W.L, Verdine, G.L. | | Deposit date: | 2005-11-26 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a DNA glycosylase searching for lesions.
Science, 311, 2006
|
|
1FN7
 
 | | COUPLING OF DAMAGE RECOGNITION AND CATALYSIS BY A HUMAN BASE-EXCISION DNA REPAIR PROTEIN | | Descriptor: | 8-OXOGUANINE DNA GLYCOSYLASE 1, CALCIUM ION, DNA (5'-D(*GP*CP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*TP*AP*CP*C)-3'), ... | | Authors: | Norman, D.P.G, Bruner, S.D, Verdine, G.L. | | Deposit date: | 2000-08-21 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Coupling of substrate recognition and catalysis by a human base-excision DNA repair protein.
J.Am.Chem.Soc., 123, 2001
|
|
2I5W
 
 | |
2H6N
 
 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me2 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-31 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2H68
 
 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-30 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2H6K
 
 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-31 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2H6Q
 
 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me3 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1DCT
 
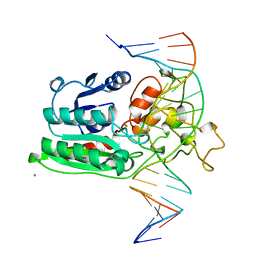 | | DNA (CYTOSINE-5) METHYLASE FROM HAEIII COVALENTLY BOUND TO DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*CP*AP*GP*CP*AP*GP*GP*(C49)P*CP*AP*CP*CP*AP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*TP*GP*GP*TP*GP*GP*(C5M)P*CP*TP*GP*CP*TP*GP*G)-3'), ... | | Authors: | Reinisch, K.M, Chen, L, Verdine, G.L, Lipscomb, W.N. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing.
Cell(Cambridge,Mass.), 82, 1995
|
|
1A66
 
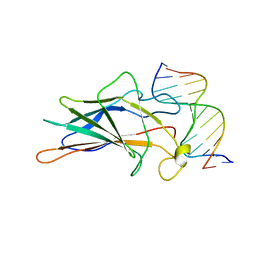 | | SOLUTION NMR STRUCTURE OF THE CORE NFATC1/DNA COMPLEX, 18 STRUCTURES | | Descriptor: | CORE NFATC1, DNA (5'-D(*CP*AP*AP*TP*TP*TP*TP*CP*CP*TP*CP*G)-3'), DNA (5'-D(*CP*GP*AP*GP*GP*AP*AP*AP*AP*TP*TP*G)-3') | | Authors: | Zhou, P, Sun, L.J, Doetsch, V, Wagner, G, Verdine, G.L. | | Deposit date: | 1998-03-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the core NFATC1/DNA complex.
Cell(Cambridge,Mass.), 92, 1998
|
|
1BNK
 
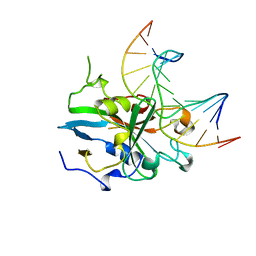 | | HUMAN 3-METHYLADENINE DNA GLYCOSYLASE COMPLEXED TO DNA | | Descriptor: | DNA (5'-D(*GP*AP*CP*AP*TP*GP*YRRP*TP*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), PROTEIN (3-METHYLADENINE DNA GLYCOSYLASE) | | Authors: | Lau, A.Y, Schaerer, O.D, Samson, L, Verdine, G.L, Ellenberger, T. | | Deposit date: | 1998-07-29 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision.
Cell(Cambridge,Mass.), 95, 1998
|
|
1A3Q
 
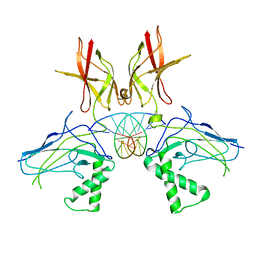 | | HUMAN NF-KAPPA-B P52 BOUND TO DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*AP*AP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*AP*TP*TP*CP*CP*CP*C)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B P52) | | Authors: | Cramer, P, Larson, C.J, Verdine, G.L, Muller, C.W. | | Deposit date: | 1998-01-23 | | Release date: | 1998-06-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the human NF-kappaB p52 homodimer-DNA complex at 2.1 A resolution.
EMBO J., 16, 1997
|
|
2NOZ
 
 | | Structure of Q315F human 8-oxoguanine glycosylase distal crosslink to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*G*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOF
 
 | | Structure of Q315F human 8-oxoguanine glycosylase proximal crosslink to 8-oxoguanine DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOI
 
 | | Structure of G42A human 8-oxoguanine glycosylase crosslinked to undamaged G-containing DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*GP*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOB
 
 | | Structure of catalytically inactive H270A human 8-oxoguanine glycosylase crosslinked to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*T*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOE
 
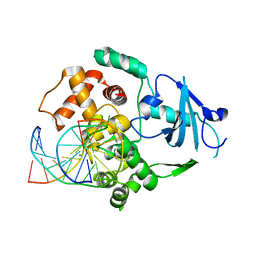 | | Structure of catalytically inactive G42A human 8-oxoguanine glycosylase complexed to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*G*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOL
 
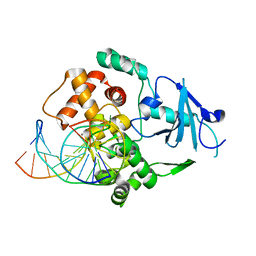 | | Structure of catalytically inactive human 8-oxoguanine glycosylase distal crosslink to oxoG DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Banerjee, A, Radom, C.T, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOH
 
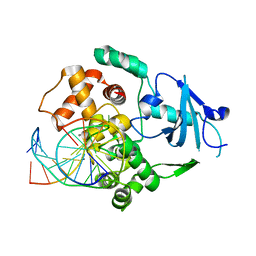 | | Structure of catalytically inactive Q315A human 8-oxoguanine glycosylase complexed to 8-oxoguanine DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
3OH9
 
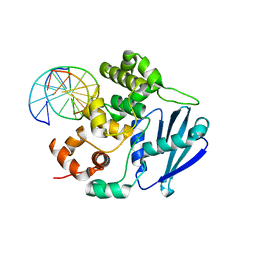 | | AlkA Undamaged DNA Complex: Interrogation of a T:A base pair | | Descriptor: | 5'-D(*AP*CP*AP*(BRU)P*GP*AP*AP*(BRU)P*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*TP*TP*CP*AP*TP*GP*T)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of Escherichia coli AlkA in Complex with Undamaged DNA.
J.Biol.Chem., 285, 2010
|
|
2CNX
 
 | | WDR5 and Histone H3 Lysine 4 dimethyl complex at 2.1 angstrom | | Descriptor: | HISTONE H3 DIMETHYL-LYSINE 4, WD-REPEAT PROTEIN 5 | | Authors: | Ruthenburg, A.J, Wang, W, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histone H3 Recognition and Presentation by the Wdr5 Module of the Mll1 Complex
Nat.Struct.Mol.Biol., 13, 2006
|
|
