7LMS
 
 | |
5FWK
 
 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
5FWP
 
 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-18 | | Release date: | 2016-10-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Atomic Structure of Hsp90:Cdc37:Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
5FWL
 
 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-18 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
5FWM
 
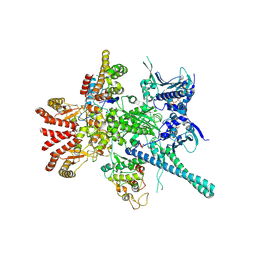 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
8UDR
 
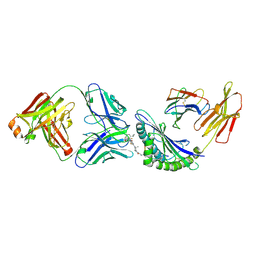 | | Structure of the P1B7 antibody bound to the Sotorasib-modified KRas G12C peptide presented by the A*03:01 MHC I complex | | Descriptor: | AMG 510 (bound form), Beta-2-microglobulin, GTPase KRas, ... | | Authors: | Chan, L.M, Roweder, P.J, Craik, C.S, Verba, K.A. | | Deposit date: | 2023-09-28 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Therapeutic targeting and structural characterization of a Sotorasib-haptenated KRAS G12C MHC I complex
To Be Published
|
|
8VSU
 
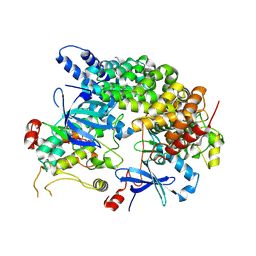 | | Cryo-EM structure of LKB1-STRADalpha-MO25alpha heterocomplex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Calcium-binding protein 39, Isoform 3 of STE20-related kinase adapter protein alpha, ... | | Authors: | Chan, L.M, Courteau, B.J, Verba, K.A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | High-resolution single-particle imaging at 100-200 keV with the Gatan Alpine direct electron detector.
J.Struct.Biol., 216, 2024
|
|
8DP5
 
 | | Structure of the PEAK3/14-3-3 complex | | Descriptor: | 14-3-3 protein beta/alpha, 14-3-3 protein epsilon, Protein PEAK3, ... | | Authors: | Torosyan, H, Paul, M, Jura, N, Verba, K.A. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into regulation of the PEAK3 pseudokinase scaffold by 14-3-3.
Nat Commun, 14, 2023
|
|
8DS6
 
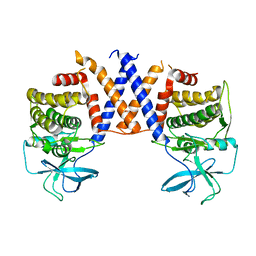 | |
8DV1
 
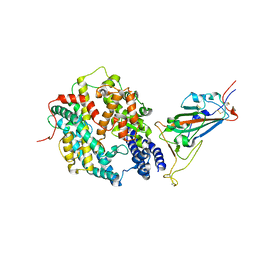 | | SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to linker variant of affinity matured ACE2 mimetic CVD432 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Immunoglobulin gamma-1 heavy chain fusion,Immunoglobulin gamma-1 heavy chain, Spike glycoprotein | | Authors: | QCRG Structural Biology Consortium, Remesh, S.G, Merz, G.E, Brilot, A.F, Chio, U, Verba, K.A. | | Deposit date: | 2022-07-27 | | Release date: | 2022-08-31 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Computational pipeline provides mechanistic understanding of Omicron variant of concern neutralizing engineered ACE2 receptor traps.
Structure, 31, 2023
|
|
8DV2
 
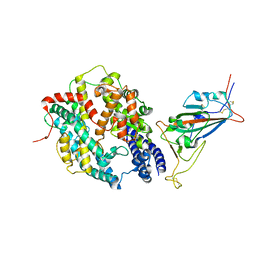 | | SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to computationally engineered ACE2 mimetic CVD293 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Immunoglobulin gamma-1 heavy chain fusion, Spike glycoprotein | | Authors: | QCRG Structural Biology Consortium, Remesh, S.G, Merz, G.E, Brilot, A.F, Chio, U, Verba, K.A. | | Deposit date: | 2022-07-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Computational pipeline provides mechanistic understanding of Omicron variant of concern neutralizing engineered ACE2 receptor traps.
Structure, 31, 2023
|
|
7MN8
 
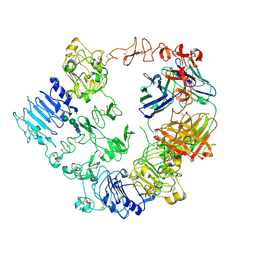 | | Structure of the HER2/HER3/NRG1b Heterodimer Extracellular Domain bound to Trastuzumab Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
7MN6
 
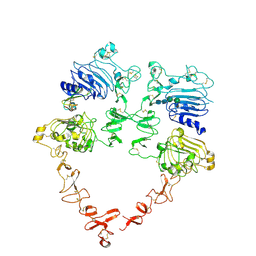 | | Structure of the HER2 S310F/HER3/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-10-27 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
7MN5
 
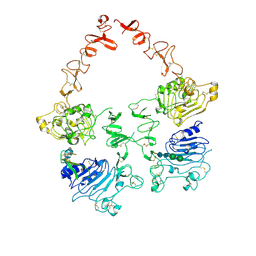 | | Structure of the HER2/HER3/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-10-27 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
7KOD
 
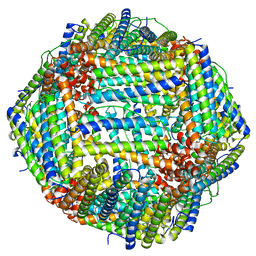 | | Cryo-EM structure of heavy chain mouse apoferritin | | Descriptor: | Ferritin heavy chain | | Authors: | Sun, M, Azumaya, C, Tse, E, Frost, A, Southworth, D, Verba, K.A, Cheng, Y, Agard, D.A. | | Deposit date: | 2020-11-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.655 Å) | | Cite: | Practical considerations for using K3 cameras in CDS mode for high-resolution and high-throughput single particle cryo-EM.
J.Struct.Biol., 213, 2021
|
|
8SMK
 
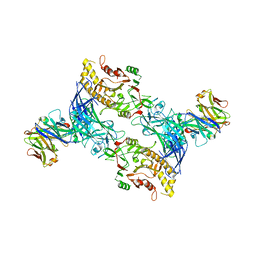 | | hPAD4 bound to Activating Fab hA362 | | Descriptor: | Activating Fab 362 heavy chain, Activating Fab 362 light chain, CALCIUM ION, ... | | Authors: | Maker, A, Verba, K.A. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4.
Nat.Chem.Biol., 20, 2024
|
|
8SML
 
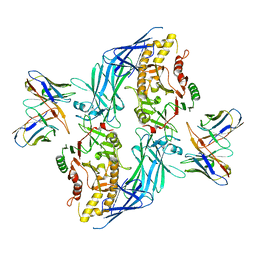 | | hPAD4 bound to inhibitory Fab hI365 | | Descriptor: | CALCIUM ION, Fab hI365 heavy chain, Fab hI365 light chain, ... | | Authors: | Maker, A, Verba, K.A. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4.
Nat.Chem.Biol., 20, 2024
|
|
8U4K
 
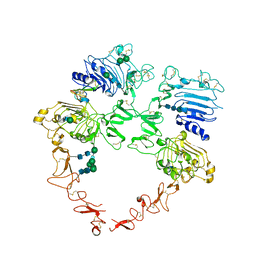 | | Structure of the HER2/HER4/BTC Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betacellulin, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
8U4I
 
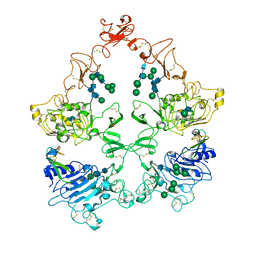 | | Structure of the HER4/NRG1b Homodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
8U4L
 
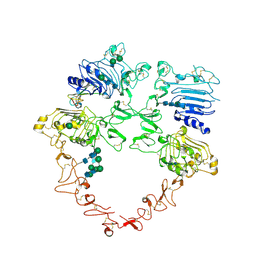 | | Structure of the HER2/HER4/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
8U4J
 
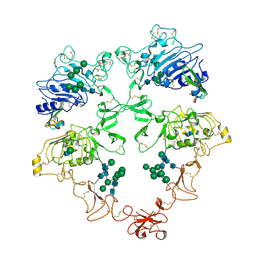 | | Structure of the HER4/BTC Homodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betacellulin, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
