1IMI
 
 | | SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 | | Descriptor: | PROTEIN (ALPHA-CONOTOXIN IMI) | | Authors: | Maslennikov, I.V, Shenkarev, Z.O, Zhmak, M.N, Tsetlin, V.I, Ivanov, V.T, Arseniev, A.S. | | Deposit date: | 1998-11-27 | | Release date: | 1999-04-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR spatial structure of alpha-conotoxin ImI reveals a common scaffold in snail and snake toxins recognizing neuronal nicotinic acetylcholine receptors.
FEBS Lett., 444, 1999
|
|
5KMW
 
 | | TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin complex | | Descriptor: | Beta-lactamase Toho-1, OPEN FORM - PENICILLIN G, PENICILLIN G, ... | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Ginell, S.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-03-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin complex
to be published
|
|
5L74
 
 | | Plexin A2 extracellular segment domains 4-5 (PSI2-IPT2), resolution 1.36 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Plexin-A2, ... | | Authors: | Kong, Y, Janssen, B.J.C, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L7N
 
 | | Plexin A1 extracellular fragment, domains 7-10 (IPT3-IPT6) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kong, Y, Janssen, B.J.C, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L56
 
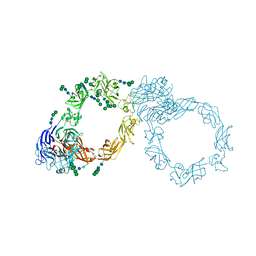 | | Plexin A1 full extracellular region, domains 1 to 10, to 4 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5C
 
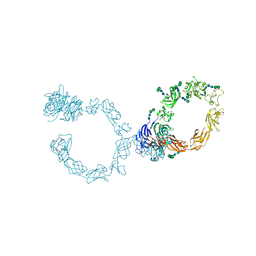 | | Plexin A1 full extracellular region, domains 1 to 10, to 6 angstrom, spacegroup P4(3)2(1)2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1, ... | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
6D4L
 
 | |
6D54
 
 | |
1SLS
 
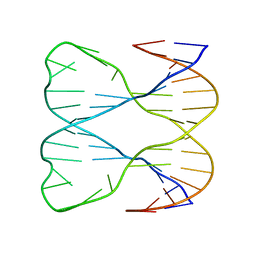 | | IMMOBILE SLIPPED-LOOP STRUCTURE (SLS) OF DNA HOMODIMER IN SOLUTION, NMR, 9 STRUCTURES | | Descriptor: | OLIGODEOXYRIBONUCLEOTIDE | | Authors: | Ulyanov, N.B, Ivanov, V.I, Minyat, E.E, Khomyakova, E.B, Petrova, M.V, Lesiak, K, James, T.L. | | Deposit date: | 1997-09-30 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A pseudosquare knot structure of DNA in solution.
Biochemistry, 37, 1998
|
|
5L5N
 
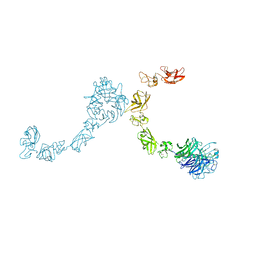 | | Plexin A4 full extracellular region, domains 1 to 7 modeled, data to 8.5 angstrom, spacegroup P4(3)22 | | Descriptor: | Plexin-A4 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.502 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5G
 
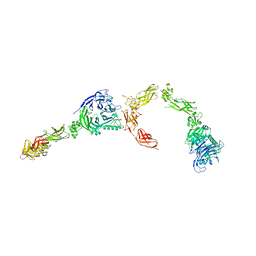 | | Plexin A2 full extracellular region, domains 1 to 8 modeled, data to 10 angstrom | | Descriptor: | Plexin-A2 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (10 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5M
 
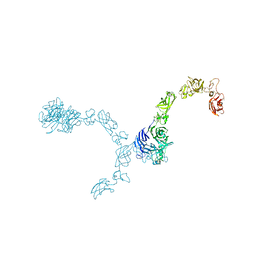 | | Plexin A4 full extracellular region, domains 1 to 7 modeled, data to 8 angstrom, spacegroup P4(3)2(1)2 | | Descriptor: | Plexin-A4 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5L
 
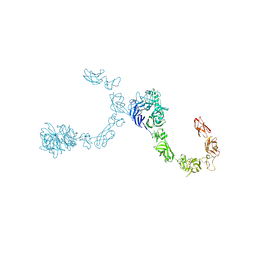 | | Plexin A4 full extracellular region, domains 1 to 8 modeled, data to 8 angstrom, spacegroup P2(1) | | Descriptor: | Plexin-A4 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.001 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
7U5O
 
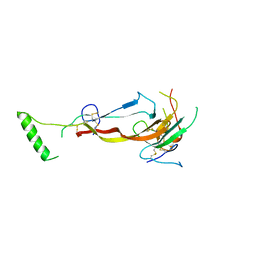 | | CRYSTAL STRUCTURE OF THE BONE MORPHOGENETIC PROTEIN RECEPTOR TYPE 2 LIGAND BINDING DOMAIN IN COMPLEX WITH ACTIVIN-B | | Descriptor: | Bone morphogenetic protein receptor type-2, Inhibin beta B chain | | Authors: | Chu, K.Y, Malik, A, Thamilselvan, V, Martinez-Hackert, E. | | Deposit date: | 2022-03-02 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Type II BMP and activin receptors BMPR2 and ACVR2A share a conserved mode of growth factor recognition.
J.Biol.Chem., 298, 2022
|
|
7U5P
 
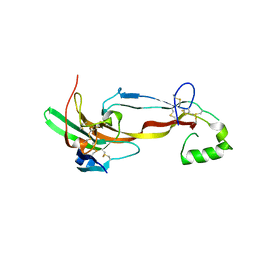 | | CRYSTAL STRUCTURE OF THE ACTIVIN RECEPTOR TYPE-2A LIGAND BINDING DOMAIN IN COMPLEX WITH ACTIVIN-A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2A, Inhibin beta A chain | | Authors: | Chu, K.Y, Malik, A, Thamilselvan, V, Martinez-Hackert, E. | | Deposit date: | 2022-03-02 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Type II BMP and activin receptors BMPR2 and ACVR2A share a conserved mode of growth factor recognition.
J.Biol.Chem., 298, 2022
|
|
6C7A
 
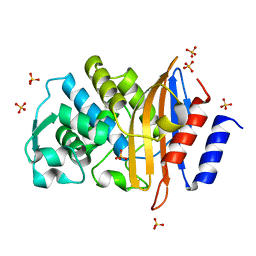 | | Conformational Changes in a Class A Beta lactamase that Prime it for Catalysis | | Descriptor: | Beta-lactamase Toho-1, SULFATE ION | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M. | | Deposit date: | 2018-01-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
6C78
 
 | | Substrate Binding Induces Conformational Changes In A Class A Beta Lactamase That Primes It For Catalysis | | Descriptor: | Beta-lactamase Toho-1 | | Authors: | Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M, Coates, L. | | Deposit date: | 2018-01-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
6C79
 
 | | Conformational Changes in a Class A Beta lactamase that Prime it for Catalysis | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase Toho-1, SULFATE ION | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M. | | Deposit date: | 2018-01-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
1X35
 
 | | Recombinant T=3 capsid of a site specific mutant of SeMV CP | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Sangita, V, Lokesh, G.L, Satheshkumar, P.S, Saravanan, V, Vijay, C.S, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2005-04-29 | | Release date: | 2005-10-18 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural studies on recombinant T = 3 capsids of Sesbania mosaic virus coat protein mutants.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1X33
 
 | | T=3 recombinant capsid of SeMV CP | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Sangita, V, Lokesh, G.L, Satheshkumar, P.S, Saravanan, V, Vijay, C.S, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2005-04-29 | | Release date: | 2005-10-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural studies on recombinant T = 3 capsids of Sesbania mosaic virus coat protein mutants.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1VAK
 
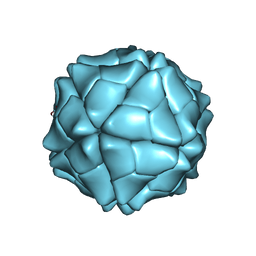 | | T=1 capsid structure of Sesbania mosaic virus coat protein deletion mutant CP-N(delta)65 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Sangita, V, Lokesh, G.L, Satheshkumer, P.S, Vijay, C.S, Saravanan, V, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2004-02-18 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | T=1 capsid structures of Sesbania mosaic virus coat protein mutants: determinants of T=3 and T=1 capsid assembly
J.Mol.Biol., 342, 2004
|
|
1VB4
 
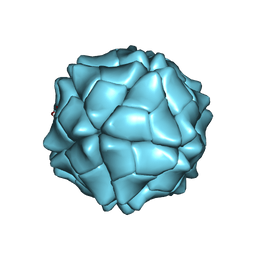 | | T=1 capsid structure of Sesbania mosaic virus coat protein deletion mutant CP-N(delta)36 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Sangita, V, Lokesh, G.L, Satheshkumar, P.S, Vijay, C.S, Saravanan, V, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2004-02-21 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | T=1 capsid structures of Sesbania mosaic virus coat protein mutants: determinants of T=3 and T=1 capsid assembly
J.Mol.Biol., 342, 2004
|
|
1VB2
 
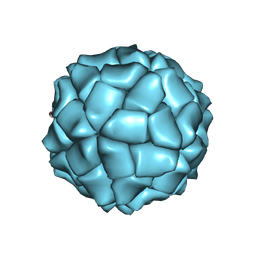 | | T=1 capsid structure of Sesbania mosaic virus coat protein deletion mutant CP-N(delta)65-D146N-D149N | | Descriptor: | coat protein | | Authors: | Sangita, V, Lokesh, G.L, Satheshkumar, P.S, Vijay, C.S, Saravanan, V, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2004-02-21 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | T=1 capsid structures of Sesbania mosaic virus coat protein mutants: determinants of T=3 and T=1 capsid assembly
J.Mol.Biol., 342, 2004
|
|
1T3Q
 
 | | Crystal structure of quinoline 2-Oxidoreductase from Pseudomonas Putida 86 | | Descriptor: | DIOXOSULFIDOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bonin, I, Martins, B.M, Purvanov, V, Fetzner, S, Huber, R, Dobbek, H. | | Deposit date: | 2004-04-27 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Active site geometry and substrate recognition of the molybdenum hydroxylase quinoline 2-oxidoreductase.
STRUCTURE, 12, 2004
|
|
