5A4G
 
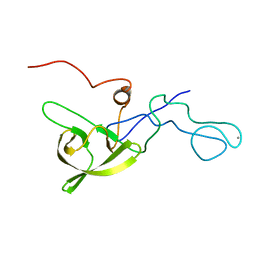 | | NMR structure of a 180 residue construct encompassing the N-terminal metal-binding site and the membrane proximal domain of SilB from Cupriavidus metallidurans CH34 | | Descriptor: | SILB, SILVER EFFLUX PROTEIN, MFP COMPONENT OF THE THREE COMPONENTS PROTON ANTIPORTER METAL EFFLUX SYSTEM, ... | | Authors: | Bersch, B, Urbina Fernandez, P, Vandenbussche, G. | | Deposit date: | 2015-06-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Investigation of the Ag+/Cu+-Binding Domains of the Periplasmic Adaptor Protein Silb from Cupriavidus Metallidurans Ch34.
Biochemistry, 55, 2016
|
|
3LJR
 
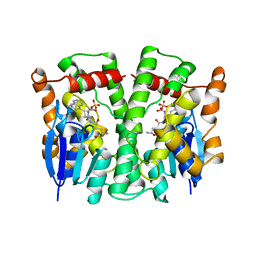 | | GLUTATHIONE TRANSFERASE (THETA CLASS) FROM HUMAN IN COMPLEX WITH THE GLUTATHIONE CONJUGATE OF 1-MENAPHTHYL SULFATE | | Descriptor: | 1-MENAPHTHYL GLUTATHIONE CONJUGATE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Rossjohn, J, Mckinstry, W.J, Oakley, A.J, Verger, D, Flanagan, J, Chelvanayagam, G, Tan, K.L, Board, P.G, Parker, M.W. | | Deposit date: | 1998-03-08 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Human theta class glutathione transferase: the crystal structure reveals a sulfate-binding pocket within a buried active site.
Structure, 6, 1998
|
|
1Y6R
 
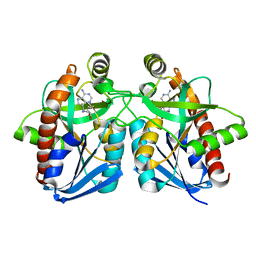 | | Crystal structure of MTA/AdoHcy nucleosidase complexed with MT-ImmA. | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
1Y6Q
 
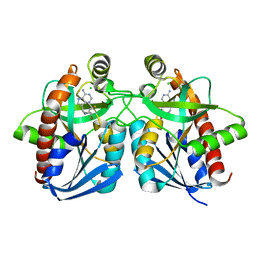 | | Cyrstal structure of MTA/AdoHcy nucleosidase complexed with MT-DADMe-ImmA | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, CHLORIDE ION, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
211D
 
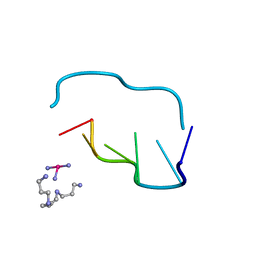 | | THE CRYSTAL AND MOLECULAR STRUCTURE OF A NEW Z-DNA CRYSTAL FORM: D[CGT(2-NH2-A) CG] AND ITS PLATINATED DERIVATIVE | | Descriptor: | DNA (5'-D(*CP*GP*TP*(1AP)P*CP*(PT(NH3)3)G)-3'), PLATINUM TRIAMINE ION, SPERMINE | | Authors: | Parkinson, G.N, Arvantis, G.M, Lessinger, L, Ginell, S.L, Jones, R, Gaffney, B, Berman, H.M. | | Deposit date: | 1995-06-13 | | Release date: | 1996-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal and molecular structure of a new Z-DNA crystal form: d[CGT(2-NH2-A)CG] and its platinated derivative.
Biochemistry, 34, 1995
|
|
210D
 
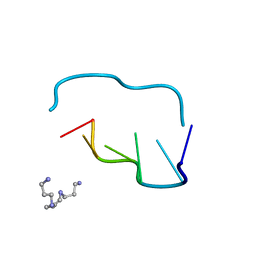 | | CRYSTAL AND MOLECULAR STRUCTURE OF A NEW Z-DNA CRYSTAL FORM: D[CGT(2-NH2-A)CG] AND ITS PLATINATED DERIVATIVE | | Descriptor: | DNA (5'-D(*CP*GP*TP*(1AP)P*CP*G)-3'), SPERMINE | | Authors: | Parkinson, G.N, Arvantis, G.M, Lessinger, L, Ginell, S.L, Jones, R, Gaffney, B, Berman, H.M. | | Deposit date: | 1995-06-13 | | Release date: | 1996-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal and molecular structure of a new Z-DNA crystal form: d[CGT(2-NH2-A)CG] and its platinated derivative.
Biochemistry, 34, 1995
|
|
3VBR
 
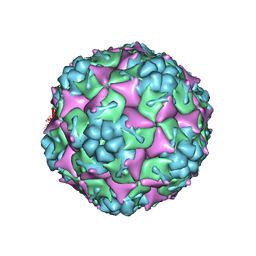 | | Crystal structure of formaldehyde treated empty human Enterovirus 71 particle (room temperature) | | Descriptor: | Genome Polyprotein, capsid protein VP0, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1SRV
 
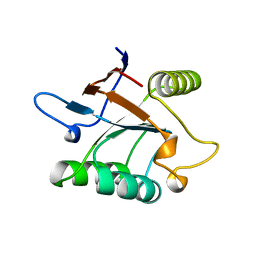 | | THERMUS THERMOPHILUS GROEL (HSP60 CLASS) FRAGMENT (APICAL DOMAIN) COMPRISING RESIDUES 192-336 | | Descriptor: | PROTEIN (GROEL (HSP60 CLASS)) | | Authors: | Walsh, M.A, Dementieva, I, Evans, G, Sanishvili, R, Joachimiak, A. | | Deposit date: | 1999-03-02 | | Release date: | 1999-03-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Taking MAD to the extreme: ultrafast protein structure determination.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3VBS
 
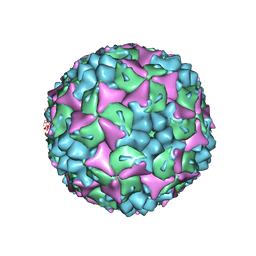 | | Crystal structure of human Enterovirus 71 | | Descriptor: | Genome Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VBU
 
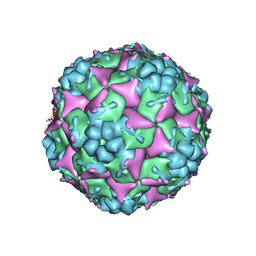 | | Crystal structure of empty human Enterovirus 71 particle | | Descriptor: | Genome Polyprotein, capsid protein VP0, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4K0E
 
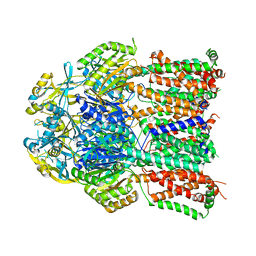 | | X-ray crystal structure of a heavy metal efflux pump, crystal form II | | Descriptor: | Heavy metal cation tricomponent efflux pump ZneA(CzcA-like), ZINC ION | | Authors: | Pak, J.E, Ngonlong Ekende, E, Vandenbussche, G, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2013-04-03 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.713 Å) | | Cite: | Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K0J
 
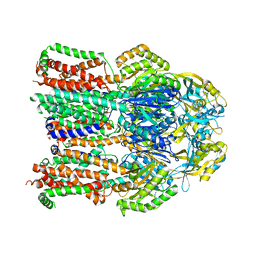 | | X-ray crystal structure of a heavy metal efflux pump, crystal form I | | Descriptor: | Heavy metal cation tricomponent efflux pump ZneA(CzcA-like), ZINC ION | | Authors: | Pak, J.E, Stroud, R.M, Ngonlong Ekende, E, Vandenbussche, G, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2013-04-04 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2BP2
 
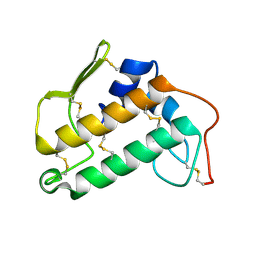 | | THE STRUCTURE OF BOVINE PANCREATIC PROPHOSPHOLIPASE A2 AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | PHOSPHOLIPASE A2 | | Authors: | Dijkstra, B.W, Vannes, G.J.H, Kalk, K.H, Brandenburg, N.P, Hol, W.G.J, Drenth, J. | | Deposit date: | 1981-06-05 | | Release date: | 1981-07-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Bovine Pancreatic Prophospholipase A2 at 3.0 Angstroms Resolution
Acta Crystallogr.,Sect.B, 38, 1982
|
|
3CRX
 
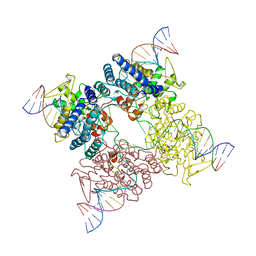 | |
3LJP
 
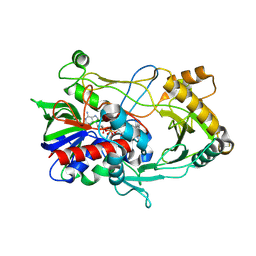 | | Crystal structure of choline oxidase V464A mutant | | Descriptor: | Choline oxidase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Finnegan, S, Agniswamy, J, Weber, I.T, Giovanni, G. | | Deposit date: | 2010-01-26 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of valine 464 in the flavin oxidation reaction catalyzed by choline oxidase.
Biochemistry, 49, 2010
|
|
1XQH
 
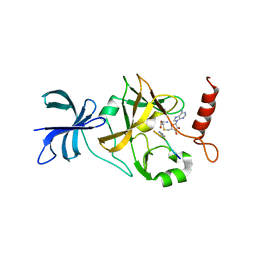 | | Crystal structure of a ternary complex of the methyltransferase SET9 (also known as SET7/9) with a P53 peptide and SAH | | Descriptor: | 9-mer peptide from tumor protein p53, Histone-lysine N-methyltransferase, H3 lysine-4 specific, ... | | Authors: | Chuikov, S, Kurash, J.K, Wilson, J.R, Xiao, B, Justin, N, Ivanov, G.S, McKinney, K, Tempst, P, Prives, C, Gamblin, S.J, Barlev, N.A, Reinberg, D. | | Deposit date: | 2004-10-12 | | Release date: | 2004-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Regulation of p53 activity through lysine methylation
Nature, 432, 2004
|
|
2CRX
 
 | |
2LJR
 
 | | GLUTATHIONE TRANSFERASE APO-FORM FROM HUMAN | | Descriptor: | GLUTATHIONE S-TRANSFERASE | | Authors: | Rossjohn, J, Mckinstry, W.J, Oakley, A.J, Verger, D, Flanagan, J, Chelvanayagam, G, Tan, K.L, Board, P.G, Parker, M.W. | | Deposit date: | 1998-03-08 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human theta class glutathione transferase: the crystal structure reveals a sulfate-binding pocket within a buried active site.
Structure, 6, 1998
|
|
3LNN
 
 | | Crystal structure of ZneB from Cupriavidus metallidurans | | Descriptor: | Membrane fusion protein (MFP) heavy metal cation efflux ZneB (CzcB-like), ZINC ION | | Authors: | Lee, J.K, De Angelis, F, Miercke, L.J, Stroud, R.M, Vandenbussche, G, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-02-02 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Metal-induced conformational changes in ZneB suggest an active role of membrane fusion proteins in efflux resistance systems.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1ZFJ
 
 | | INOSINE MONOPHOSPHATE DEHYDROGENASE (IMPDH; EC 1.1.1.205) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | INOSINE MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID | | Authors: | Zhang, R, Evans, G, Rotella, F.J, Westbrook, E.M, Beno, D, Huberman, E, Joachimiak, A, Collart, F.R. | | Deposit date: | 1999-03-29 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characteristics and crystal structure of bacterial inosine-5'-monophosphate dehydrogenase.
Biochemistry, 38, 1999
|
|
2PPN
 
 | |
2L55
 
 | |
2PPP
 
 | |
