7GER
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-f7918075-5 (Mpro-x11499) | | Descriptor: | 2-(3-chlorophenyl)-N-(2,6-naphthyridin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GCJ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-590ac91e-19 (Mpro-x10626) | | Descriptor: | (1R,6S,7r)-N-(4-methylpyridin-3-yl)bicyclo[4.1.0]heptane-7-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GES
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-bb423b95-7 (Mpro-x11501) | | Descriptor: | 2-(3-chlorophenyl)-N-(isoquinolin-4-yl)-N-methylacetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GCM
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with NIR-THE-c331be7a-2 (Mpro-x10678) | | Descriptor: | 3-[(4R)-2-acetyl-1,2,3,4-tetrahydroisoquinolin-4-yl]benzonitrile, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GF4
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-590ac91e-17 (Mpro-x11560) | | Descriptor: | (2S)-2-(difluoromethoxy)-N-(4-methylpyridin-3-yl)propanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2PGL
 
 | | Catalysis associated conformational changes revealed by human CD38 complexed with a non-hydrolyzable substrate analog | | Descriptor: | ADP-ribosyl cyclase 1, N1-CYCLIC INOSINE 5'-DIPHOSPHORIBOSE | | Authors: | Liu, Q, Kriksunov, I.A, Moreau, C, Graeff, R, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2007-04-09 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Catalysis-associated Conformational Changes Revealed by Human CD38 Complexed with a Non-hydrolyzable Substrate Analog
J.Biol.Chem., 282, 2007
|
|
3T5N
 
 | | 1.8A crystal structure of Lassa virus nucleoprotein in complex with ssRNA | | Descriptor: | NICKEL (II) ION, Nucleoprotein, RNA (5'-R(P*UP*AP*UP*CP*UP*C)-3') | | Authors: | Hastie, K.M, Liu, T, King, L.B, Ngo, N, Zandonatti, M.A, Woods, V.L, de la Torre, J.C, Saphire, E.O. | | Deposit date: | 2011-07-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3T5Q
 
 | | 3A structure of Lassa virus nucleoprotein in complex with ssRNA | | Descriptor: | Nucleoprotein, PHOSPHATE ION, RNA (5'-R(P*UP*AP*UP*CP*UP*C)-3'), ... | | Authors: | Hastie, K.M, Liu, T, King, L.B, Ngo, N, Zandonatti, M.A, Woods, V.L, de la Torre, J.C, Saphire, E.O. | | Deposit date: | 2011-07-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6MCD
 
 | | Crystal Structure of tris-thiolate Pb(II) complex with adjacent water in a de novo Three Stranded Coiled Coil Peptide | | Descriptor: | LEAD (II) ION, Pb(II)(GRAND Coil Ser L12CL16A)-, ZINC ION | | Authors: | Tolbert, A.E, Ruckthong, L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2018-08-31 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Heteromeric three-stranded coiled coils designed using a Pb(II)(Cys)3template mediated strategy.
Nat.Chem., 12, 2020
|
|
2PGJ
 
 | | Catalysis associated conformational changes revealed by human cd38 complexed with a non-hydrolyzable substrate analog | | Descriptor: | ADP-ribosyl cyclase 1, N1-CYCLIC INOSINE 5'-DIPHOSPHORIBOSE | | Authors: | Liu, Q, Kriksunov, I.A, Moreau, C, Graeff, R, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2007-04-09 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Catalysis-associated Conformational Changes Revealed by Human CD38 Complexed with a Non-hydrolyzable Substrate Analog
J.Biol.Chem., 282, 2007
|
|
4ZTH
 
 | | Structure of human p38aMAPK-arylpyridazinylpyridine fragment complex used in inhibitor discovery | | Descriptor: | 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, 6-chloro-3-phenyl-4-(pyridin-4-yl)pyridazine, BETA-MERCAPTOETHANOL, ... | | Authors: | Grum-Tokars, V.L, Roy, S.M, Watterson, D.M. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Selective and Brain Penetrant p38 alpha MAPK Inhibitor Candidate for Neurologic and Neuropsychiatric Disorders That Attenuates Neuroinflammation and Cognitive Dysfunction.
J.Med.Chem., 62, 2019
|
|
6XTJ
 
 | | The high resolution structure of the FERM domain of human FERMT2 | | Descriptor: | CITRIC ACID, Fermitin family homolog 2,Fermitin family homolog 2,Fermitin family homolog 2 | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The high resolution structure of the FERM domain of human FERMT2
To Be Published
|
|
5A7I
 
 | | Crystal structure of INPP5B in complex with biphenyl 3,3',4,4',5,5'- hexakisphosphate | | Descriptor: | Biphenyl 3,3',4,4',5,5'-hexakisphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Mills, S.J, Silvander, C, Cozier, G, Potter, B.V.L, Norldund, P. | | Deposit date: | 2015-07-06 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal Structures of Type-II Inositol Polyphosphate 5-Phosphatase Inpp5B with Synthetic Inositol Polyphosphate Surrogates Reveal New Mechanistic Insights for the Inositol 5-Phosphatase Family.
Biochemistry, 55, 2016
|
|
6WS1
 
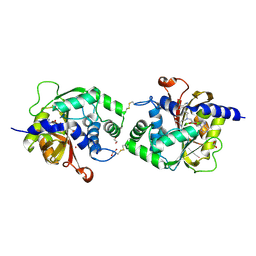 | | Crystal structure of human phenylethanolamine N-methyltransferase (PNMT) in complex with (2S)-2-amino-4-((((2R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)(3-(7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl)propyl)amino)butanoic acid and AdoHcy (SAH) | | Descriptor: | 1,2-ETHANEDIOL, 5'-([(3S)-3-amino-3-carboxypropyl]{3-[(4R)-7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl]propyl}amino)-5'-deoxyadenosine, CADMIUM ION, ... | | Authors: | Harijan, R.K, Mahmoodi, N, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2020-04-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Transition-State Analogues of PhenylethanolamineN-Methyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6O1T
 
 | | BOVINE SALIVARY PROTEIN FORM 30B WITH OLEIC ACID | | Descriptor: | CALCIUM ION, OLEIC ACID, Short palate, ... | | Authors: | Zhang, H, Arcus, V.L. | | Deposit date: | 2019-02-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three dimensional structure of Bovine Salivary Protein 30b (BSP30b) and its interaction with specific rumen bacteria.
Plos One, 14, 2019
|
|
5A7J
 
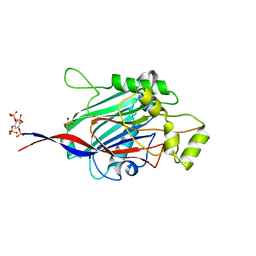 | | Crystal structure of INPP5B in complex with benzene 1,2,4,5- tetrakisphosphate | | Descriptor: | (2,4,5-triphosphonooxyphenyl) dihydrogen phosphate, GLYCEROL, SULFATE ION, ... | | Authors: | Tresaugues, L, Mills, S.J, Silvander, C, Cozier, G, Potter, B.V.L, Nordlund, P. | | Deposit date: | 2015-07-06 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Type-II Inositol Polyphosphate 5-Phosphatase Inpp5B with Synthetic Inositol Polyphosphate Surrogates Reveal New Mechanistic Insights for the Inositol 5-Phosphatase Family.
Biochemistry, 55, 2016
|
|
5WCZ
 
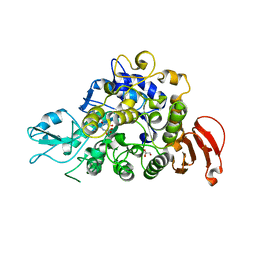 | |
5WD6
 
 | | bovine salivary protein form 30b | | Descriptor: | CALCIUM ION, Short palate, lung and nasal epithelium carcinoma-associated protein 2B | | Authors: | Zhang, H, Arcus, V.L. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-11 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three dimensional structure of Bovine Salivary Protein 30b (BSP30b) and its interaction with specific rumen bacteria.
Plos One, 14, 2019
|
|
5E57
 
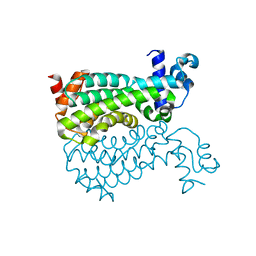 | | Crystal structure of Mycobacterium smegmatis AmtR | | Descriptor: | ACETATE ION, Transcription regulator AmtR | | Authors: | Vickers, C.J, McKenzie, J.L, Arcus, V.L. | | Deposit date: | 2015-10-07 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Function of AmtR in Mycobacterium smegmatis: Implications for Post-Transcriptional Regulation of Urea Metabolism through a Small Antisense RNA.
J.Mol.Biol., 428, 2016
|
|
4ECE
 
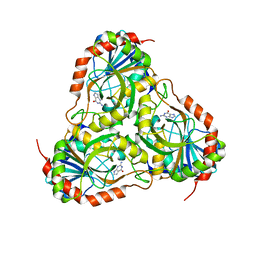 | | Crystal structure of purine nucleoside phosphorylase (W16Y, W94Y, W178Y, H257W) mutant from human complexed with guanine | | Descriptor: | GUANINE, Purine nucleoside phosphorylase | | Authors: | Haapalainen, A.M, Ho, M.C, Suarez, J.J, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-03-26 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalytic Site Conformations in Human PNP by (19)F-NMR and Crystallography.
Chem.Biol., 20, 2013
|
|
1G2P
 
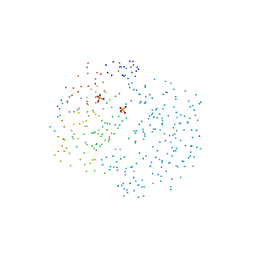 | | CRYSTAL STRUCTURE OF ADENINE PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | ADENINE PHOSPHORIBOSYLTRANSFERASE 1, SULFATE ION | | Authors: | Shi, W, Tanaka, K.S.E, Almo, S.C, Schramm, V.L. | | Deposit date: | 2000-10-20 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of adenine phosphoribosyltransferase from Saccharomyces cerevisiae.
Biochemistry, 40, 2001
|
|
4EB8
 
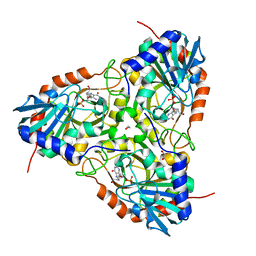 | | Crystal structure of purine nucleoside phosphorylase (W16Y, W94Y, W178Y, H257W) mutant from human complexed with DADMe-ImmG and phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, ... | | Authors: | Ho, M.C, Haapalainen, A.M, Suarez, J.J, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-03-23 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic Site Conformations in Human PNP by (19)F-NMR and Crystallography.
Chem.Biol., 20, 2013
|
|
1G81
 
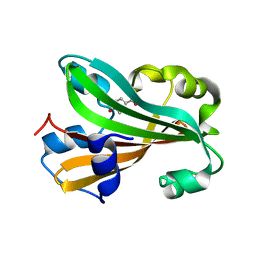 | | CHORISMATE LYASE WITH BOUND PRODUCT, ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | CHORISMATE LYASE, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M.J, Vilker, V.L, Howard, A. | | Deposit date: | 2000-11-15 | | Release date: | 2001-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The crystal structure of chorismate lyase shows a new fold and a tightly retained product.
Proteins, 44, 2001
|
|
8BLU
 
 | | The PDZ domains of human SDCBP | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Daniel-Mozo, M, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The PDZ domains of human SDCBP
To Be Published
|
|
8BNT
 
 | | The DH domain of ARHGEF2 bound to RhoA | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, Rho guanine nucleotide exchange factor 2, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Grosjean, H, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2022-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The DH domain of ARHGEF2 bound to RhoA
To Be Published
|
|
