5XDW
 
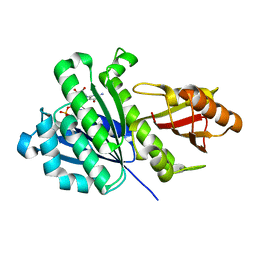 | | Staphylococcus aureus FtsZ 12-316 G196S | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Maeda, Y, Mizohata, E, Inoue, T, Kaul, M, Parhi, A.K, LaVoie, E.J, Pilch, D.S, Matsumura, H. | | Deposit date: | 2017-03-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Flexibility of an Inhibitor Overcomes Drug Resistance Mutations in Staphylococcus aureus FtsZ
ACS Chem. Biol., 12, 2017
|
|
1IDZ
 
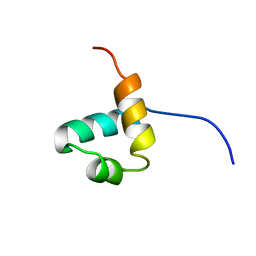 | |
5YCH
 
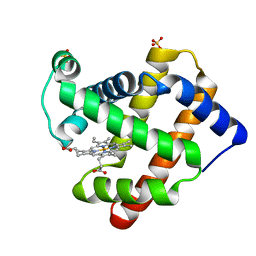 | | Ancestral myoglobin aMbWb of Basilosaurus relative (monophyly) | | Descriptor: | Ancestral myoglobin aMbWb of Basilosaurus relative (monophyly), PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Nakagawa, T, Tsuneshige, A, Shirai, T. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.354 Å) | | Cite: | Tracing whale myoglobin evolution by resurrecting ancient proteins.
Sci Rep, 8, 2018
|
|
3VHI
 
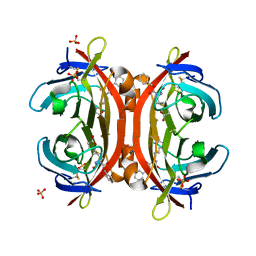 | | Crystal structure of monoZ-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-{(3aS,4S,6aR)-1-[(benzyloxy)carbonyl]-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl}pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
3VI2
 
 | | Crystal Structure Analysis of Plasmodium falciparum OMP Decarboxylase in complex with inhibitor HMOA | | Descriptor: | 4-(2-hydroxy-4-methoxyphenyl)-4-oxobutanoic acid, Orotidine 5'-phosphate decarboxylase, SODIUM ION | | Authors: | Takashima, Y, Mizohata, E, Krungkrai, S.R, Matsumura, H, Krungkrai, J, Horii, T, Inoue, T. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The in silico screening and X-ray structure analysis of the inhibitor complex of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase
J.Biochem., 152, 2012
|
|
5YCG
 
 | | Ancestral myoglobin aMbWp of Pakicetus relative | | Descriptor: | Ancestral myoglobin aMbWp of Pakicetus relative, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Nakagawa, T, Tsuneshige, A, Shirai, T. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tracing whale myoglobin evolution by resurrecting ancient proteins.
Sci Rep, 8, 2018
|
|
3VHM
 
 | | Crystal structure of NPC-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4R,6aR)-1-{[(1R)-1-(6-nitro-1,3-benzodioxol-5-yl)ethoxy]carbonyl}-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-29 | | Release date: | 2011-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
1HSR
 
 | | BINDING MODE OF BENZHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BENZHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1997-07-01 | | Release date: | 1998-07-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding mode of benzhydroxamic acid to Arthromyces ramosus peroxidase shown by X-ray crystallographic analysis of the complex at 1.6 A resolution.
FEBS Lett., 412, 1997
|
|
5YCI
 
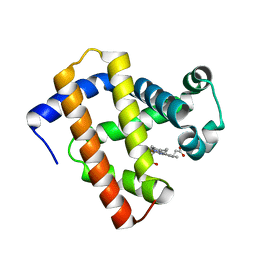 | | Ancestral myoglobin aMbWb' of Basilosaurus relative (polyphyly) | | Descriptor: | Ancestral myoglobin aMbWb' of Basilosaurus relative (polyphyly), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Nakagawa, T, Tsuneshige, A, Shirai, T. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Tracing whale myoglobin evolution by resurrecting ancient proteins.
Sci Rep, 8, 2018
|
|
5YCE
 
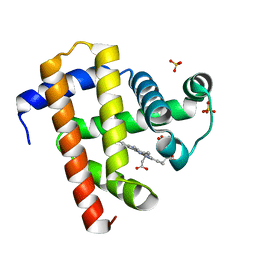 | | Sperm whale myoglobin swMb | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Nakagawa, T, Tsuneshige, A, Shirai, T. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Tracing whale myoglobin evolution by resurrecting ancient proteins.
Sci Rep, 8, 2018
|
|
3WT1
 
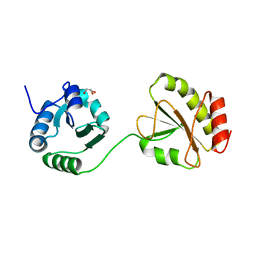 | | Crystal structure of the b'-a' domain of thermophilic fungal protein disulfide isomerase (reduced form) | | Descriptor: | GLYCEROL, Protein disulfide-isomerase | | Authors: | Inagaki, K, Satoh, T, Itoh, S.G, Okumura, H, Kato, K. | | Deposit date: | 2014-04-02 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Redox-dependent conformational transition of catalytic domain of protein disulfide isomerase indicated by crystal structure-based molecular dynamics simulation
Chem.Phys.Lett., 618, 2015
|
|
3WQU
 
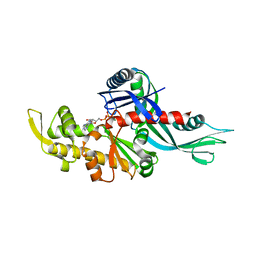 | | Staphylococcus aureus FtsA complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, MAGNESIUM ION | | Authors: | Fujita, J, Maeda, Y, Miyazaki, Y, Inoue, T, Matsumura, H. | | Deposit date: | 2014-02-01 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of FtsA from Staphylococcus aureus
FEBS Lett., 588, 2014
|
|
2D0B
 
 | | Crystal structure of Bst-RNase HIII in complex with Mg2+ | | Descriptor: | MAGNESIUM ION, ribonuclease HIII | | Authors: | Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2005-07-31 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus
J.Mol.Biol., 356, 2006
|
|
2D0A
 
 | | Crystal structure of Bst-RNase HIII | | Descriptor: | ribonuclease HIII | | Authors: | Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2005-07-31 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus
J.Mol.Biol., 356, 2006
|
|
3VHH
 
 | | Crystal structure of DiMe-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4S,6aR)-1,3-dimethyl-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
3WQO
 
 | | Crystal structure of D-tagatose 3-epimerase-like protein | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein MJ1311 | | Authors: | Uechi, K, Takata, G, Yoneda, K, Ohshima, T, Sakuraba, H. | | Deposit date: | 2014-01-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of D-tagatose 3-epimerase-like protein from Methanocaldococcus jannaschii
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1SU4
 
 | | Crystal structure of calcium ATPase with two bound calcium ions | | Descriptor: | CALCIUM ION, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | Authors: | Toyoshima, C, Nakasako, M, Nomura, H, Ogawa, H. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution
Nature, 405, 2000
|
|
5XVH
 
 | | Crystal structure of the NADP+ and tartrate-bound complex of L-serine 3-dehydrogenase from the hyperthermophilic archaeon Pyrobaculum calidifontis | | Descriptor: | 6-phosphogluconate dehydrogenase, NAD-binding protein, ACETIC ACID, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2017-06-28 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of the NADP+and tartrate-bound complex of L-serine 3-dehydrogenase from the hyperthermophilic archaeon Pyrobaculum calidifontis.
Extremophiles, 22, 2018
|
|
1V9L
 
 | | L-glutamate dehydrogenase from Pyrobaculum islandicum complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, glutamate dehydrogenase | | Authors: | Bhuiya, M.W, Sakuraba, H, Ohshima, T, Imagawa, T, Katunuma, N, Tsuge, H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The First Crystal Structure of Hyperthermostable NAD-dependent Glutamate Dehydrogenase from Pyrobaculum islandicum
J.Mol.Biol., 345, 2005
|
|
3VGW
 
 | | Crystal structure of monoAc-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4S,6aR)-1-acetyl-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-21 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
1LAW
 
 | |
2CWH
 
 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae complexed with NADPH and pyrrole-2-carboxylate | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRROLE-2-CARBOXYLATE, delta1-piperideine-2-carboxylate reductase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2005-06-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
3WQT
 
 | | Staphylococcus aureus FtsA complexed with AMPPNP | | Descriptor: | CHLORIDE ION, Cell division protein FtsA, MAGNESIUM ION, ... | | Authors: | Fujita, J, Maeda, Y, Miyazaki, Y, Inoue, T, Matsumura, H. | | Deposit date: | 2014-02-01 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FtsA from Staphylococcus aureus
FEBS Lett., 588, 2014
|
|
1LAV
 
 | |
1IDY
 
 | |
