5H5G
 
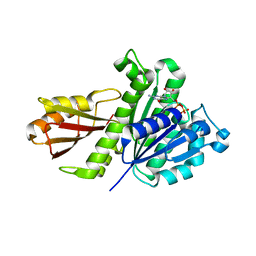 | | Staphylococcus aureus FtsZ-GDP in T and R states | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
1PKF
 
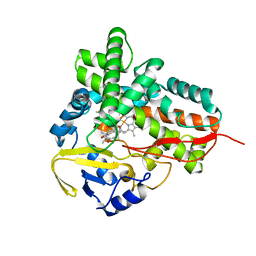 | | Crystal Structure of Epothilone D-bound Cytochrome P450epoK | | Descriptor: | EPOTHILONE D, PROTOPORPHYRIN IX CONTAINING FE, cytochrome p450EpoK | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-06-05 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Epothilone D-bound, Epothilone B-bound, and Substrate-free Forms of Cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
5H5I
 
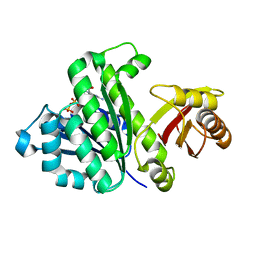 | | Staphylococcus aureus FtsZ-GDP R29A mutant in R state | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
1Q5D
 
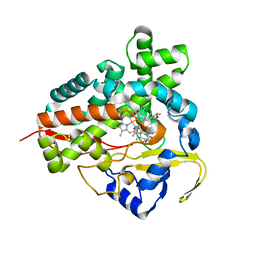 | | Epothilone B-bound Cytochrome P450epoK | | Descriptor: | 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-THIAZOL-4-YL)VINYL]-4,17-DIOXABICYCLO[14.1.0]HEPTADECANE-5,9-DIONE, P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
1Q5E
 
 | | Substrate-free Cytochrome P450epoK | | Descriptor: | P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
3WG7
 
 | | A 1.9 angstrom radiation damage free X-ray structure of large (420KDa) protein by femtosecond crystallography | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Hirata, K, Shinzawa-Itoh, K, Yano, N, Takemura, S, Kato, K, Hatanaka, M, Muramoto, K, Kawahara, T, Tsukihara, T, Yamashita, E, Tono, K, Ueno, G, Hikima, T, Murakami, H, Inubushi, Y, Yabashi, M, Ishikawa, T, Yamamoto, M, Ogura, T, Sugimoto, H, Shen, J.R, Yoshikawa, S, Ago, H. | | Deposit date: | 2013-07-29 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL.
Nat.Methods, 11, 2014
|
|
1IWA
 
 | | RUBISCO FROM GALDIERIA PARTITA | | Descriptor: | SULFATE ION, ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit, ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit | | Authors: | Okano, Y, Mizohata, E, Xie, Y, Matsumura, H, Sugawara, H, Inoue, T, Yokota, A, Kai, Y. | | Deposit date: | 2002-04-30 | | Release date: | 2003-04-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure of Galdieria Rubisco Complexed with one sulfate ion per active site
FEBS LETT., 527, 2002
|
|
2YVS
 
 | |
7XVX
 
 | |
5Y5F
 
 | | Structure of cytochrome P450nor in NO-bound state: damaged by low-dose (0.72 MGy) X-ray | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
3W0K
 
 | |
5Y5G
 
 | | Structure of cytochrome P450nor in NO-bound state: damaged by high-dose (5.7 MGy) X-ray | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
7W7C
 
 | | Heme exporter in the unliganded form | | Descriptor: | Putative ABC transport system integral membrane protein, Putative ABC transport system, ATP-binding protein, ... | | Authors: | Rahman, M.M, Hisano, T, Nakamura, H, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4O6S
 
 | | 1.32A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant, Zinc Bound) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
7ZCS
 
 | | Nitrite-bound MSOX movie series dataset 65 (52 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - water ligand (final) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCP
 
 | | Nitrite-bound MSOX movie series dataset 17 (13.6 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - nitric oxide (NO) intermediate | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCR
 
 | | Nitrite-bound MSOX movie series dataset 38 (30.4 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - water ligand | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCO
 
 | | Nitrite-bound MSOX movie series dataset 8 (6.4 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - nitrite/NO intermediate | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCQ
 
 | | Nitrite-bound MSOX movie series dataset 25 (20 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - NO/water intermediate | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCN
 
 | | Nitrite-bound MSOX movie series dataset 1 (0.8 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - nitrite (start) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3ETA
 
 | | Kinase domain of insulin receptor complexed with a pyrrolo pyridine inhibitor | | Descriptor: | 1-(3-{5-[4-(aminomethyl)phenyl]-1H-pyrrolo[2,3-b]pyridin-3-yl}phenyl)-3-(2-phenoxyphenyl)urea, insulin receptor, kinase domain | | Authors: | Patnaik, S, Stevens, K, Gerding, R, Deanda, F, Shotwell, B, Tang, J, Hamajima, T, Nakamura, H, Leesnitzer, A, Hassell, A, Shewchuk, L, Kumar, R, Lei, H, Chamberlain, S. | | Deposit date: | 2008-10-07 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 3,5-disubstituted-1H-pyrrolo[2,3-b]pyridines as potent inhibitors of the insulin-like growth factor-1 receptor (IGF-1R) tyrosine kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3WUQ
 
 | | Structure of the entire stalk region of the dynein motor domain | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1 | | Authors: | Nishikawa, Y, Oyama, T, Kamiya, N, Kon, T, Toyoshima, Y.Y, Nakamura, H, Kurisu, G. | | Deposit date: | 2014-05-01 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the entire stalk region of the Dynein motor domain
J.Mol.Biol., 426, 2014
|
|
1QM8
 
 | | Structure of Bacteriorhodopsin at 100 K | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | Authors: | Takeda, K, Matsui, Y, Sato, H, Hino, T, Kanamori, E, Okumura, H, Yamane, T, Kamiya, N, Kouyama, T. | | Deposit date: | 1999-09-22 | | Release date: | 2000-08-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Three-Dimensional Crystal of Bacteriorhodopsin Obtained by Successive Fusion of the Vesicular Assemblies.
J.Mol.Biol., 283, 1998
|
|
1WTJ
 
 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae pvar.tomato | | Descriptor: | ureidoglycolate dehydrogenase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2004-11-24 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
1WMJ
 
 | |
