6BKC
 
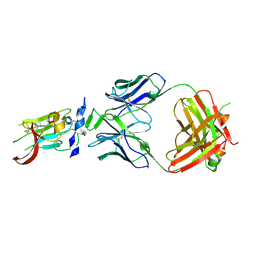 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody AR3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab AR3B heavy chain, Fab AR3B light chain, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2017-11-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies.
Sci Adv, 5, 2019
|
|
6BZV
 
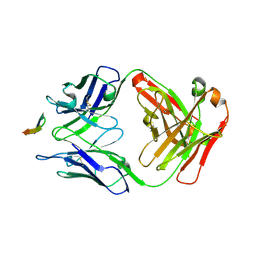 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the GL precursor of the broadly neutralizing antibody 19B3 | | Descriptor: | 19B3 GL Heavy Chain, 19B3 GL Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BKD
 
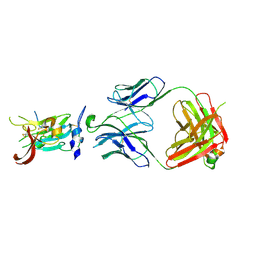 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody AR3D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab AR3D heavy chain, Fab AR3D light chain, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2017-11-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies.
Sci Adv, 5, 2019
|
|
6BZU
 
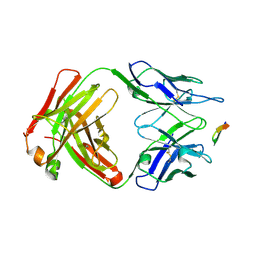 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody 19B3 | | Descriptor: | 19B3 Heavy Chain, 19B3 Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BZY
 
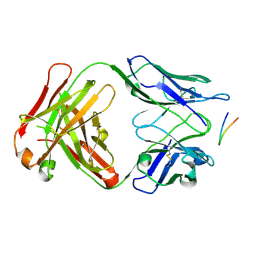 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the 22D11 broadly neutralizing antibody | | Descriptor: | 22D11 Heavy Chain, 22D11 Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BZW
 
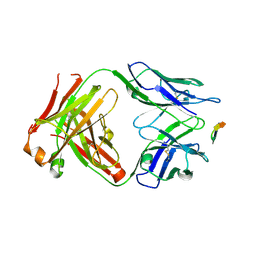 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the GL precursor of the broadly neutralizing antibody AP33 | | Descriptor: | AP33 GL Heavy Chain, AP33 GL Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5TH0
 
 | |
5THC
 
 | |
5TGO
 
 | |
5TGU
 
 | |
5TH1
 
 | |
5THF
 
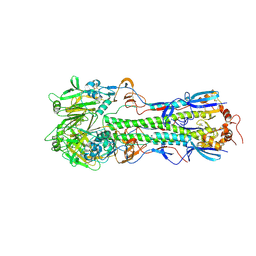 | |
5THB
 
 | |
3OD6
 
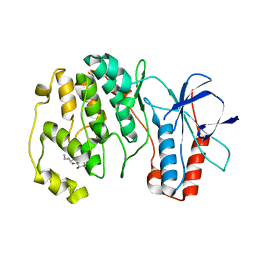 | | Crystal structure of p38alpha Y323T active mutant | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Livnah, O, Tzarum, N. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.682 Å) | | Cite: | Active mutants of the TCR-mediated p38alpha alternative activation site show changes in the phosphorylation lip and DEF site formation.
J.Mol.Biol., 405, 2011
|
|
4S34
 
 | | ERK2 (I84A) in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Livnah, O, Karamansha, Y, Tzarum, N. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multiple mechanisms render Erk proteins MEK-independent
To be Published
|
|
4S33
 
 | | ERK2 R65S mutant complexed with AMP-PNP | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Livnah, O, Karamansha, Y, Tzarum, N. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Multiple mechanisms render Erk proteins MEK-independent
To be Published
|
|
3OEF
 
 | | Crystal structure of Y323F inactive mutant of p38alpha MAP kinase | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Livnah, O, Tzarum, N. | | Deposit date: | 2010-08-12 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active mutants of the TCR-mediated p38alpha alternative activation site show changes in the phosphorylation lip and DEF site formation.
J.Mol.Biol., 405, 2011
|
|
3ODY
 
 | | Crystal structure of p38alpha Y323Q active mutant | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Livnah, O, Tzarum, N. | | Deposit date: | 2010-08-12 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active mutants of the TCR-mediated p38alpha alternative activation site show changes in the phosphorylation lip and DEF site formation.
J.Mol.Biol., 405, 2011
|
|
3ODZ
 
 | | Crystal structure of P38alpha Y323R active mutant | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Livnah, O, Tzarum, N. | | Deposit date: | 2010-08-12 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active mutants of the TCR-mediated p38alpha alternative activation site show changes in the phosphorylation lip and DEF site formation.
J.Mol.Biol., 405, 2011
|
|
4GEO
 
 | | P38a MAP kinase DEF-pocket penta mutant (M194A, L195A, H228A, I229A, Y258A) | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Livnah, O, Tzarum, N. | | Deposit date: | 2012-08-02 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | P38a MAP kinase DEF-pocket penta mutant (M194A, L195A, H228A, I229A, Y258A)
To be Published
|
|
4E6A
 
 | | p38a-PIA23 complex | | Descriptor: | (2S)-2-methoxy-3-(octadecyloxy)propyl (1R,2R,3R,4S,6S)-2,3,4-trihydroxy-6-(2-methylpropoxy)cyclohexyl hydrogen (S)-phosphate, Mitogen-activated protein kinase 14 | | Authors: | Livnah, O, Tzarum, N, Eisenberg-Domovich, Y. | | Deposit date: | 2012-03-15 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Lipid Molecules Induce p38 alpha Activation via a Novel Molecular Switch.
J.Mol.Biol., 424, 2012
|
|
4E6C
 
 | | p38a-perifosine Complex | | Descriptor: | (1,1-dimethylpiperidin-1-ium-4-yl) octadecyl hydrogen phosphate, Mitogen-activated protein kinase 14 | | Authors: | Livnah, O, Tzarum, N, Eisenberg-Domovich, Y. | | Deposit date: | 2012-03-15 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Lipid Molecules Induce p38 alpha Activation via a Novel Molecular Switch.
J.Mol.Biol., 424, 2012
|
|
4E8A
 
 | | The crystal structure of p38a MAP kinase in complex with PIA24 | | Descriptor: | (1R,2S,3R,4S,6S)-6-(cyclohexylmethoxy)-2,3,4-trihydroxycyclohexyl (2R)-2-methoxy-3-(octadecyloxy)propyl hydrogen (S)-phosphate, Mitogen-activated protein kinase 14 | | Authors: | Livnah, O, Tzarum, N, Eisenberg-Domovich, Y. | | Deposit date: | 2012-03-20 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lipid Molecules Induce p38 alpha Activation via a Novel Molecular Switch.
J.Mol.Biol., 424, 2012
|
|
4E5A
 
 | |
4E5B
 
 | |
