4QSH
 
 | | Crystal Structure of L. monocytogenes Pyruvate Carboxylase in complex with Cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CITRATE ANION, MANGANESE (II) ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2014-07-04 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The Cyclic Dinucleotide c-di-AMP Is an Allosteric Regulator of Metabolic Enzyme Function.
Cell(Cambridge,Mass.), 158, 2014
|
|
4N9X
 
 | | Crystal Structure of the OCTAPRENYL-METHYL-METHOXY-BENZQ MOLECULE from Erwina carotovora subsp. atroseptica strain SCRI 1043 / ATCC BAA-672, Northeast Structural Genomics Consortium (NESG) Target EwR161 | | Descriptor: | Putative monooxygenase | | Authors: | Kuzin, A, Chen, Y, Lew, S, Seetharaman, J, Mao, L, Xiao, R, Owens, L.A, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-21 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal Structure of the OCTAPRENYL-METHYL-METHOXY-BENZQ MOLECULE from Erwina carotovora subsp. atroseptica strain SCRI 1043 / ATCC BAA-672, Northeast Structural Genomics Consortium (NESG) Target EwR161
To be Published
|
|
3G8D
 
 | | Crystal structure of the biotin carboxylase subunit, E296A mutant, of acetyl-COA carboxylase from Escherichia coli | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, MAGNESIUM ION, ... | | Authors: | Chou, C.Y, Yu, L.P, Tong, L. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism.
J.Biol.Chem., 284, 2009
|
|
1WYZ
 
 | | X-Ray structure of the putative methyltransferase from Bacteroides thetaiotaomicron VPI-5482 at the resolution 2.5 A. Norteast Structural Genomics Consortium target Btr28 | | Descriptor: | putative S-adenosylmethionine-dependent methyltransferase | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Vorobiev, S.M, Acton, T, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-21 | | Release date: | 2005-03-08 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray structure of the putative methyltransferase from Bacteroides thetaiotaomicron VPI-5482 at the resolution 2.5 A. Norteast Structural Genomics Consortium target Btr28
To be Published
|
|
5UC9
 
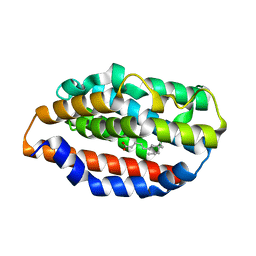 | |
1XUA
 
 | | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, Phenazine biosynthesis protein phzF | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XSQ
 
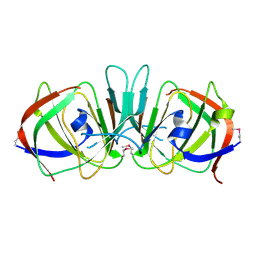 | | Crystal structure of ureidoglycolate hydrolase from E.coli. Northeast Structural Genomics Consortium target ET81. | | Descriptor: | Ureidoglycolate hydrolase | | Authors: | Kuzin, A.P, Vorobiev, S.M, Abashidze, M, Acton, T.B, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-19 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ureidoglycolate hydrolase from E.coli. Northeast Structural Genomics Consortium target ET81.
To be Published
|
|
4PY9
 
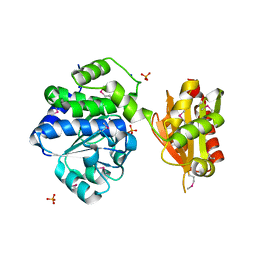 | | Crystal structure of an exopolyphosphatase-related protein from Bacteroides Fragilis. Northeast Structural Genomics target BFR192 | | Descriptor: | PHOSPHATE ION, Putative exopolyphosphatase-related protein, SODIUM ION | | Authors: | Seetharaman, J, Abashidze, M, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-03-26 | | Release date: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of an exopolyphosphatase-related protein from Bacteroides Fragilis. Northeast Structural Genomics target BFR192
To be Published
|
|
4LNY
 
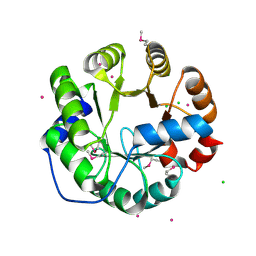 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR422 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Engineered Protein OR422 | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Crystal Structure of Engineered Protein OR422.
To be Published
|
|
5V22
 
 | |
1ZHV
 
 | | X-ray Crystal Structure Protein Atu0741 from Agobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR8. | | Descriptor: | hypothetical protein Atu0741 | | Authors: | Vorobiev, S.M, Kuzin, A, Abashidze, M, Forouhar, F, Xio, R, Ma, L.-C, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-26 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of hypothetical protein Atu071 from Agobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR8.
To be Published
|
|
1ZKD
 
 | | X-Ray structure of the putative protein Q6N1P6 from Rhodopseudomonas palustris at the resolution 2.1 A , Northeast Structural Genomics Consortium target RpR58 | | Descriptor: | DUF185 | | Authors: | Kuzin, A.P, Yong, W, Vorobiev, S.M, Acton, T, Ma, L, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-02 | | Release date: | 2005-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray structure of the putative protein Q6N1P6 from Rhodopseudomonas palustris at the resolution 2.1 A , Northeast Structural Genomics Consortium target RpR58
To be Published
|
|
2I4R
 
 | | Crystal structure of the V-type ATP synthase subunit F from Archaeoglobus fulgidus. NESG target GR52A. | | Descriptor: | V-type ATP synthase subunit F | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Zhao, L, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-08-22 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the V-type ATP synthase subunit F from Archaeoglobus fulgidus
To be Published
|
|
1BI8
 
 | | MECHANISM OF G1 CYCLIN DEPENDENT KINASE INHIBITION FROM THE STRUCTURES CDK6-P19INK4D INHIBITOR COMPLEX | | Descriptor: | CYCLIN-DEPENDENT KINASE 6, CYCLIN-DEPENDENT KINASE INHIBITOR | | Authors: | Russo, A.A, Tong, L, Lee, J.O, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1998-06-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a.
Nature, 395, 1998
|
|
1BI7
 
 | | MECHANISM OF G1 CYCLIN DEPENDENT KINASE INHIBITION FROM THE STRUCTURE OF THE CDK6-P16INK4A TUMOR SUPPRESSOR COMPLEX | | Descriptor: | CYCLIN-DEPENDENT KINASE 6, MULTIPLE TUMOR SUPPRESSOR | | Authors: | Russo, A.A, Tong, L, Lee, J.O, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1998-06-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a.
Nature, 395, 1998
|
|
1ZN6
 
 | | X-ray Crystal Structure of Protein Q7WLM8 from Bordetella bronchiseptica. Northeast Structural Genomics Consortium Target BoR19. | | Descriptor: | SULFATE ION, phage-related conserved hypothetical protein | | Authors: | Kuzin, A.P, Yong, W, Vorobiev, S.M, Xiao, R, Ma, L.-C, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-11 | | Release date: | 2005-05-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structure of the hypothetical protein Q7WLM8 from Bordetella bronchiseptica. Northeast Structural Genomics Consortium target BoR19.
To be Published
|
|
5VYZ
 
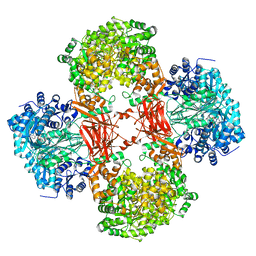 | | Crystal structure of Lactococcus lactis pyruvate carboxylase in complex with cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2017-05-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V8W
 
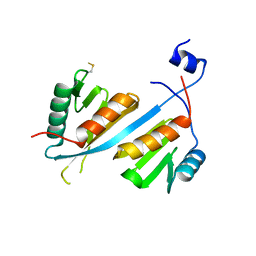 | | Crystal structure of human Integrator IntS9-IntS11 CTD complex | | Descriptor: | Integrator complex subunit 11, Integrator complex subunit 9 | | Authors: | Wu, Y, Tong, L. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-12 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the interaction between Integrator subunits IntS9 and IntS11 and its functional importance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1XW8
 
 | | X-ray structure of putative lactam utilization protein YBGL. Northeast Structural Genomics Consortium target ET90. | | Descriptor: | UPF0271 protein ybgL | | Authors: | Kuzin, A.P, Chen, Y, Vorobiev, S.M, Acton, T.B, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-29 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of putative lactam utilization protein YBGL.
Northeast Structural Genomics Consortium target ET90.
To be Published
|
|
5VZ0
 
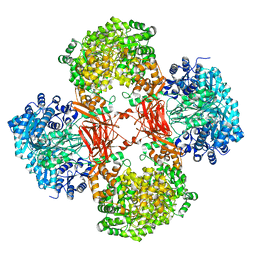 | | Crystal structure of Lactococcus lactis pyruvate carboxylase G746A mutant in complex with cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2017-05-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1XM3
 
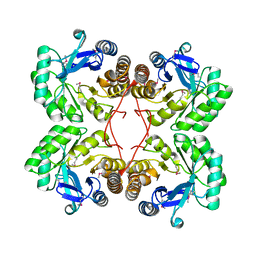 | | Crystal structure of Northeast Structural Genomics Target SR156 | | Descriptor: | Thiazole biosynthesis protein thiG | | Authors: | Kuzin, A, Abashidze, M, Vorobiev, S, Forouhar, F, Acton, T, Ma, L, Xiao, R, Montelione, G, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Northeast Structural Genomics Target SR156
To be Published
|
|
1ZQ7
 
 | | X-Ray Crystal Structure of Protein Q8PZK8 from Methanosarcina mazei. Northeast Structural Genomics Consortium Target MaR9. | | Descriptor: | Hypothetical protein MM0484 | | Authors: | Kuzin, A.P, Abashidze, M, Vorobiev, S.M, Yong, W, Xiao, R, Ma, L.-C, Acton, T, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-18 | | Release date: | 2005-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | X-Ray structure of the hypothetical protein Q8PZK8 from Methanosarcina mazei at the resolution 2.1A. Northeast Structural Genomics Consortium target MaR9.
To be Published
|
|
2A8E
 
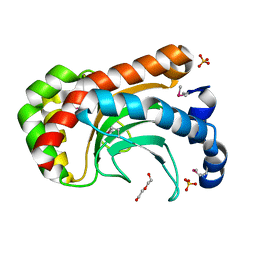 | | Three-dimensional structure of Bacillus subtilis Q45498 putative protein at resolution 2.5A. Northeast Structural Genomics Consortium target SR204. | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, hypothetical protein yktB | | Authors: | Kuzin, A.P, Su, M, Yong, W, Vorobiev, S, Acton, T, Xiao, R, Conover, K, Ma, L.-C, Cunningham, K.E, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-07-07 | | Release date: | 2005-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of Bacillus subtilis Q45498 putative protein at resolution 2.5A. Northeast Structural Genomics Consortium target SR204.
To be Published
|
|
1YDM
 
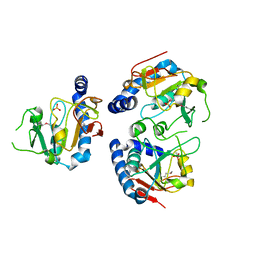 | | X-Ray structure of Northeast Structural Genomics target SR44 | | Descriptor: | Hypothetical protein yqgN, SULFATE ION | | Authors: | Kuzin, A.P, Jianwei, S, Vorobiev, S, Acton, T, Xia, R, Ma, L.-C, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-24 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray structure of Northeast Structural Genomics target SR44
To be Published
|
|
2AFC
 
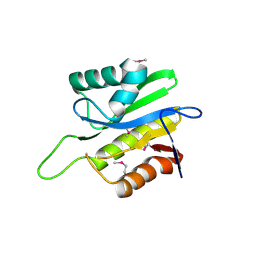 | | X-Ray Crystal Structure of Protein Q8A8B0 from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR9. | | Descriptor: | conserved hypothetical protein | | Authors: | Kuzin, A.P, Abashidze, M, Vorobiev, S.M, Acton, T, Xiao, R, Conover, K, Ma, L.-C, Cunninggham, K.E, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-07-25 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of Q8A8B0 hypothetical protein from Bacteroides thetaiotaomicron
To be Published
|
|
