7UNJ
 
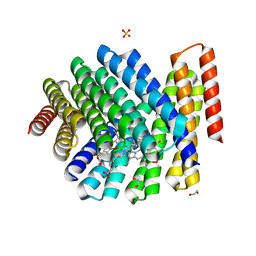 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester matching geometry of purple bacterial special pair, SP1-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, SP1-ZnPPaM designed chlorophyll dimer protein, SULFATE ION, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
7UNH
 
 | |
7UNI
 
 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester, SP2-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, SP2-ZnPPaM designed chlorophyll dimer protein, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
3KO2
 
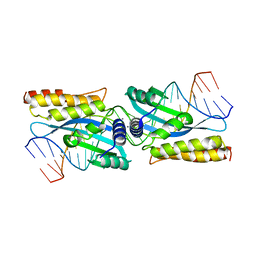 | | I-MsoI re-designed for altered DNA cleavage specificity (-7C) | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*GP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*CP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Taylor, G.K, Stoddard, B.L. | | Deposit date: | 2009-11-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
3MX4
 
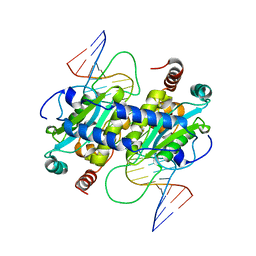 | | DNA binding and cleavage by the GIY-YIG endonuclease R.Eco29KI inactive variant E142Q | | Descriptor: | DNA (5'-D(P*CP*GP*GP*GP*AP*GP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*CP*GP*CP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*CP*GP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*CP*TP*CP*CP*CP*G)-3'), Eco29kIR | | Authors: | Mak, A.N.S, Lambert, A.R, Stoddard, B.L. | | Deposit date: | 2010-05-06 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Folding, DNA Recognition, and Function of GIY-YIG Endonucleases: Crystal Structures of R.Eco29kI.
Structure, 18, 2010
|
|
1IDE
 
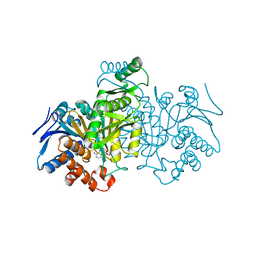 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT STEADY-STATE INTERMEDIATE COMPLEX (LAUE DETERMINATION) | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
7T8L
 
 | | BrxR from Acinetobacter BREX type I phage restriction system | | Descriptor: | 1,2-ETHANEDIOL, BrxR, CHLORIDE ION | | Authors: | Doyle, L, Kaiser, B, Stoddard, B. | | Deposit date: | 2021-12-16 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and characterization of the WYL BrxR protein and its gene as separable regulatory elements of a BREX phage restriction system.
Nucleic Acids Res., 50, 2022
|
|
8EIL
 
 | | C-Terminal Domain of BrxL from Acinetobacter BREX type I phage restriction system | | Descriptor: | MALONIC ACID, Protease Lon-related BREX system protein BrxL, SUCCINIC ACID | | Authors: | Doyle, L.A, Stoddard, B.L, Kaiser, B. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure, substrate binding and activity of a unique AAA+ protein: the BrxL phage restriction factor.
Nucleic Acids Res., 51, 2023
|
|
1AF5
 
 | | GROUP I MOBILE INTRON ENDONUCLEASE | | Descriptor: | I-CREI | | Authors: | Heath, P.J, Stephens, K.M, Monnat Junior, R.J, Stoddard, B.L. | | Deposit date: | 1997-03-21 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of I-Crel, a group I intron-encoded homing endonuclease.
Nat.Struct.Biol., 4, 1997
|
|
3E0L
 
 | | Computationally Designed Ammelide Deaminase | | Descriptor: | Guanine deaminase, ZINC ION | | Authors: | Murphy, P.M, Bolduc, J.M, Gallaher, J.L, Stoddard, B.L, Baker, D. | | Deposit date: | 2008-07-31 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Alteration of enzyme specificity by computational loop remodeling and design.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4YXY
 
 | |
5ESP
 
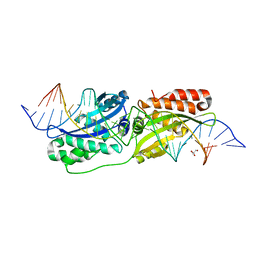 | |
3MIS
 
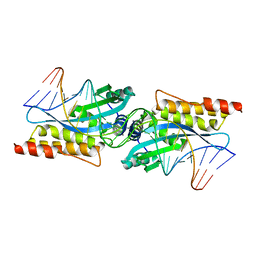 | | I-MsoI re-designed for altered DNA cleavage specificity (-8G) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*GP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*CP*TP*CP*CP*G)-3'), ... | | Authors: | Taylor, G.K, Stoddard, B.L. | | Deposit date: | 2010-04-12 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
2EX5
 
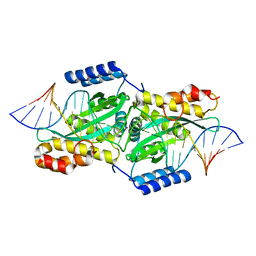 | |
7T8K
 
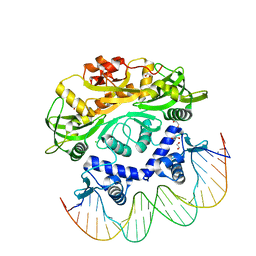 | | BrxR from Acinetobacter BREX type I phage restriction system bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, BrxR, CHLORIDE ION, ... | | Authors: | Doyle, L, Kaiser, B, Stoddard, B. | | Deposit date: | 2021-12-16 | | Release date: | 2022-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and characterization of the WYL BrxR protein and its gene as separable regulatory elements of a BREX phage restriction system.
Nucleic Acids Res., 50, 2022
|
|
6CZG
 
 | |
6CZJ
 
 | |
4R6G
 
 | |
4R5D
 
 | |
6CZI
 
 | | Structure of a redesigned beta barrel, mFAP1, bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP1 | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
4R58
 
 | |
4IWW
 
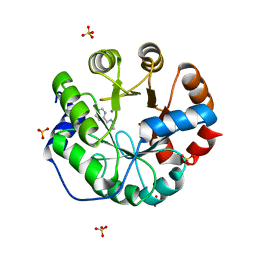 | | Computational Design of an Unnatural Amino Acid Metalloprotein with Atomic Level Accuracy | | Descriptor: | COBALT (II) ION, SULFATE ION, Unnatural Amino Acid Mediated Metalloprotein | | Authors: | Mills, J, Bolduc, J, Khare, S, Stoddard, B, Baker, D. | | Deposit date: | 2013-01-24 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of an unnatural amino Acid dependent metalloprotein with atomic level accuracy.
J.Am.Chem.Soc., 135, 2013
|
|
4R5C
 
 | |
7RDR
 
 | |
4R6F
 
 | |
