7FI9
 
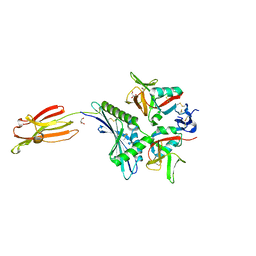 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | GLYCEROL, MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI7
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI8
 
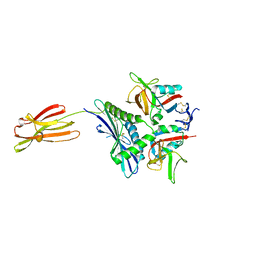 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI6
 
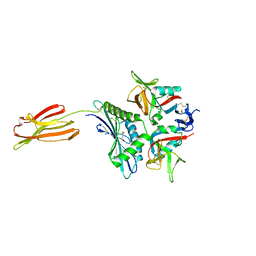 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI5
 
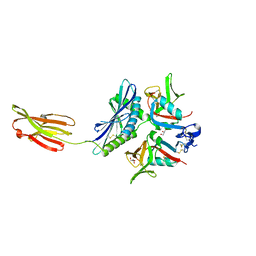 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | GLYCEROL, MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
2H3H
 
 | |
5AC3
 
 | | Crystal structure of PAM12A | | Descriptor: | ACETIC ACID, CADMIUM ION, PEPTIDE AMIDASE | | Authors: | Wu, B, Wijma, H.J, Song, L, Rozeboom, H.J, Poloni, C, Tian, Y, Arif, M.I, Nuijens, T, Quadflieg, P.J.L.M, Szymanski, W, Feringa, B.L, Janssen, D.B. | | Deposit date: | 2015-08-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Versatile Peptide C-Terminal Functionalization Via a Computationally Peptide Amidase
Acs Catalysis, 2016
|
|
4L5Q
 
 | | Crystal structure of p202 HIN1 | | Descriptor: | Interferon-activable protein 202 | | Authors: | Yin, Q, Tian, Y, Wu, H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Molecular Mechanism for p202-Mediated Specific Inhibition of AIM2 Inflammasome Activation.
Cell Rep, 4, 2013
|
|
4L5R
 
 | |
4L5S
 
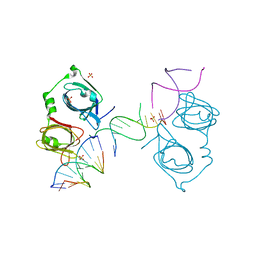 | | p202 HIN1 in complex with 12-mer dsDNA | | Descriptor: | 12-mer DNA, Interferon-activable protein 202, SULFATE ION | | Authors: | Yin, Q, Tian, Y, Wu, H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Molecular Mechanism for p202-Mediated Specific Inhibition of AIM2 Inflammasome Activation.
Cell Rep, 4, 2013
|
|
4L5T
 
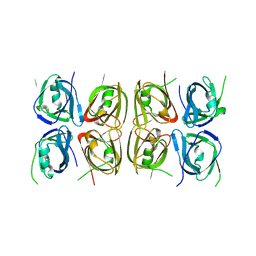 | | Crystal structure of the tetrameric p202 HIN2 | | Descriptor: | Interferon-activable protein 202 | | Authors: | Yin, Q, Tian, Y, Wu, H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.405 Å) | | Cite: | Molecular Mechanism for p202-Mediated Specific Inhibition of AIM2 Inflammasome Activation.
Cell Rep, 4, 2013
|
|
2N2L
 
 | | NMR structure of yersinia pestis ail (attachment invasion locus) in decylphosphocholine micelles calculated with implicit membrane solvation | | Descriptor: | Outer membrane protein X | | Authors: | Marassi, F.M, Ding, Y, Tian, Y, Schwieters, C.D, Yao, Y. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone structure of Yersinia pestis Ail determined in micelles by NMR-restrained simulated annealing with implicit membrane solvation.
J.Biomol.Nmr, 63, 2015
|
|
5Y3R
 
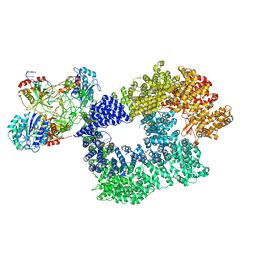 | | Cryo-EM structure of Human DNA-PK Holoenzyme | | Descriptor: | DNA (34-MER), DNA (36-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Yin, X, Liu, M, Tian, Y, Wang, J, Xu, Y. | | Deposit date: | 2017-07-29 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structure of human DNA-PK holoenzyme
Cell Res., 27, 2017
|
|
2LK9
 
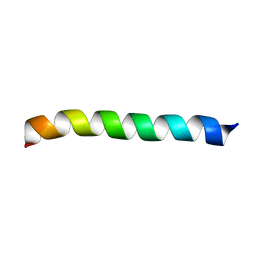 | | Structure of BST-2/Tetherin Transmembrane Domain | | Descriptor: | Bone marrow stromal antigen 2 | | Authors: | Skasko, M, Wang, Y, Tian, Y, Tokarev, A, Munguia, J, Ruiz, A, Stephens, E, Opella, S, Guatelli, J. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HIV-1 Vpu Protein Antagonizes Innate Restriction Factor BST-2 via Lipid-embedded Helix-Helix Interactions.
J.Biol.Chem., 287, 2012
|
|
5ZCS
 
 | | 4.9 Angstrom Cryo-EM structure of human mTOR complex 2 | | Descriptor: | Rapamycin-insensitive companion of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex 2 subunit MAPKAP1, ... | | Authors: | Chen, X, Liu, M, Tian, Y, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of human mTOR complex 2.
Cell Res., 28, 2018
|
|
6K9L
 
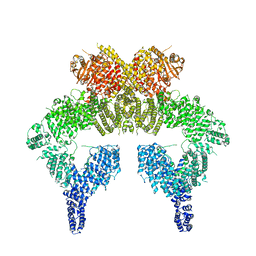 | | 4.27 Angstrom resolution cryo-EM structure of human dimeric ATM kinase | | Descriptor: | Serine-protein kinase ATM | | Authors: | Xiao, J, Liu, M, Qi, Y, Chaban, Y, Gao, C, Tian, Y, Yu, Z, Li, J, Zhang, P, Xu, Y. | | Deposit date: | 2019-06-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural insights into the activation of ATM kinase.
Cell Res., 29, 2019
|
|
5YZ0
 
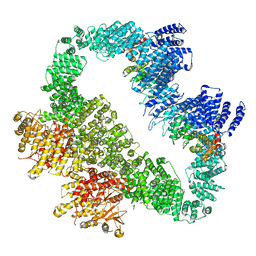 | | Cryo-EM Structure of human ATR-ATRIP complex | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR | | Authors: | Rao, Q, Liu, M, Tian, Y, Wu, Z, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-01-31 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structure of human ATR-ATRIP complex.
Cell Res., 28, 2018
|
|
4NH3
 
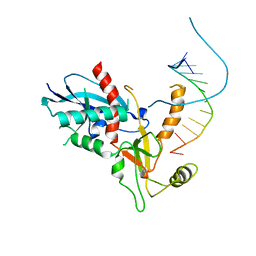 | |
4NGG
 
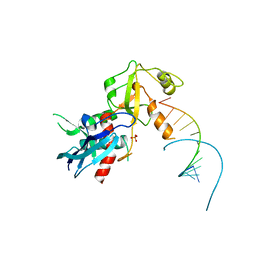 | |
4NGC
 
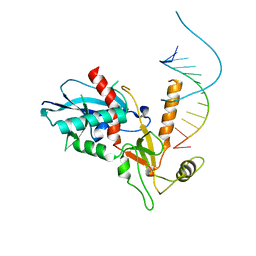 | |
4NH6
 
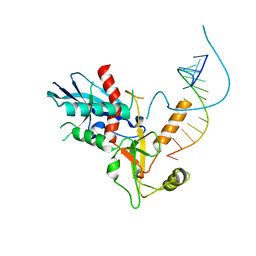 | |
4NGF
 
 | |
4NHA
 
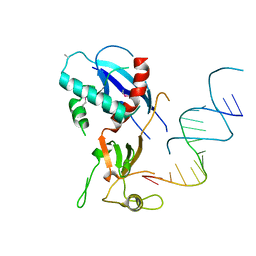 | | Structure of human Dicer Platform-PAZ-Connector Helix cassette in complex with 16-mer siRNA having 5'-p and UU-3' ends (3.4 Angstrom resolution) | | Descriptor: | 5'-R(P*GP*CP*GP*UP*UP*GP*GP*CP*CP*AP*AP*CP*GP*CP*UP*U)-3', Endoribonuclease Dicer | | Authors: | Simanshu, D.K, Tian, Y, Ma, J.-B, Patel, D.J. | | Deposit date: | 2013-11-04 | | Release date: | 2014-03-05 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | A Phosphate-Binding Pocket within the Platform-PAZ-Connector Helix Cassette of Human Dicer.
Mol.Cell, 53, 2014
|
|
4NGB
 
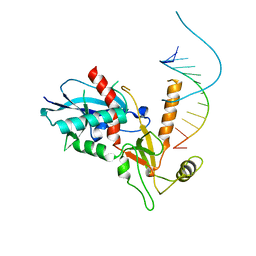 | |
7TDQ
 
 | |
