1B28
 
 | | ARC REPRESSOR MYL MUTANT FROM SALMONELLA BACTERIOPHAGE P22 | | Descriptor: | PROTEIN (REGULATORY PROTEIN ARC) | | Authors: | Rietveld, A.W.M, Nooren, I.M.A, Sauer, R.T, Kaptein, R, Boelens, R. | | Deposit date: | 1998-12-05 | | Release date: | 1999-11-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of an Arc repressor mutant reveal premelting conformational changes related to DNA binding.
Biochemistry, 38, 1999
|
|
1T3O
 
 | | Solution structure of CsrA, a bacterial carbon storage regulatory protein | | Descriptor: | Carbon storage regulator | | Authors: | Koharudin, L.M.I, Georgiou, T, Kleanthous, C, Geoffrey, R, Kaptein, R, Boelens, R. | | Deposit date: | 2004-04-27 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A model for RNA binding by the bacterial carbon storage regulatory protein, CsrA
To be Published
|
|
1B14
 
 | | Alcohol Dehydrogenase from Drosophila Lebanonensis Binary Complex with NAD+ | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Benach, J, Atrian, S, Gonzalez-Duarte, R, Ladenstein, R. | | Deposit date: | 1998-11-25 | | Release date: | 1999-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The catalytic reaction and inhibition mechanism of Drosophila alcohol dehydrogenase: observation of an enzyme-bound NAD-ketone adduct at 1.4 A resolution by X-ray crystallography.
J.Mol.Biol., 289, 1999
|
|
1LEB
 
 | | SOLUTION STRUCTURE OF THE LEXA REPRESSOR DNA BINDING DETERMINED BY 1H NMR SPECTROSCOPY | | Descriptor: | LEXA REPRESSOR DNA BINDING DOMAIN | | Authors: | Fogh, R.H, Ottleben, G, Rueterjans, H, Schnarr, M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-05-11 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy.
EMBO J., 13, 1994
|
|
3TO7
 
 | | Crystal structure of yeast Esa1 HAT domain bound to coenzyme A with active site lysine acetylated | | Descriptor: | CACODYLIC ACID, COENZYME A, GLYCEROL, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
2CKC
 
 | | Solution structures of the BRK domains of the human Chromo Helicase Domain 7 and 8, reveals structural similarity with GYF domain suggesting a role in protein interaction | | Descriptor: | CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 7 | | Authors: | Ab, E, de Jong, R.N, Diercks, T, Xiaoyun, J, Daniels, M, Kaptein, R, Folkers, G.E. | | Deposit date: | 2006-04-14 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the Brk Domains of the Human Chromo Helicase Domain 7 and 8, Reveals Structural Similarity with Gyf Domain Suggesting a Role in Protein Interaction
To be Published
|
|
1SFV
 
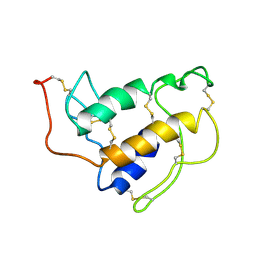 | | PORCINE PANCREAS PHOSPHOLIPASE A2, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B, Tessari, M, Boelens, R, Dijkman, R, Kaptein, R, De Haas, G.H, Verheij, H.M. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2 complexed with micelles and a competitive inhibitor.
J.Biomol.NMR, 5, 1995
|
|
1SFW
 
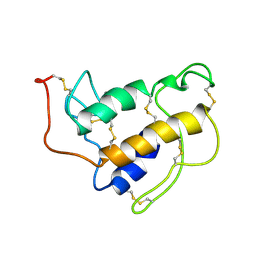 | | PORCINE PANCREAS PHOSPHOLIPASE A2, NMR, 18 STRUCTURES | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B, Tessari, M, Boelens, R, Dijkman, R, Kaptein, R, De Haas, G.H, Verheij, H.M. | | Deposit date: | 1996-02-23 | | Release date: | 1996-07-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2 complexed with micelles and a competitive inhibitor.
J.Biomol.NMR, 5, 1995
|
|
3CO7
 
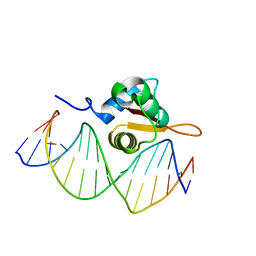 | | Crystal Structure of FoxO1 DBD Bound to DBE2 DNA | | Descriptor: | DNA (5'-D(*DCP*DAP*DAP*DAP*DAP*DTP*DGP*DTP*DAP*DAP*DAP*DCP*DAP*DAP*DGP*DA)-3'), DNA (5'-D(*DTP*DCP*DTP*DTP*DGP*DTP*DTP*DTP*DAP*DCP*DAP*DTP*DTP*DTP*DTP*DG)-3'), Forkhead box protein O1 | | Authors: | Brent, M.M, Anand, R, Marmorstein, R. | | Deposit date: | 2008-03-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Basis for DNA Recognition by FoxO1 and Its Regulation by Posttranslational Modification.
Structure, 16, 2008
|
|
1CM0
 
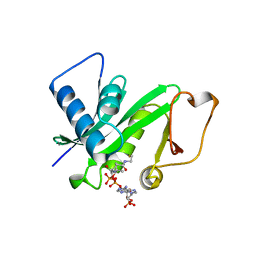 | | CRYSTAL STRUCTURE OF THE PCAF/COENZYME-A COMPLEX | | Descriptor: | COENZYME A, P300/CBP ASSOCIATING FACTOR | | Authors: | Clements, A, Rojas, J.R, Trievel, R.C, Wang, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-12 | | Release date: | 1999-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A.
EMBO J., 18, 1999
|
|
3CO6
 
 | | Crystal Structure of FoxO1 DBD Bound to DBE1 DNA | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DCP*DAP*DAP*DGP*DGP*DTP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DCP*DCP*DA)-3'), ... | | Authors: | Brent, M.M, Anand, R, Marmorstein, R. | | Deposit date: | 2008-03-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for DNA Recognition by FoxO1 and Its Regulation by Posttranslational Modification.
Structure, 16, 2008
|
|
3COA
 
 | | Crystal Structure of FoxO1 DBD Bound to IRE DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*DCP*DAP*DAP*DGP*DCP*DAP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DCP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DGP*DTP*DTP*DTP*DGP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DG)-3'), ... | | Authors: | Brent, M.M, Anand, R, Marmorstein, R. | | Deposit date: | 2008-03-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for DNA Recognition by FoxO1 and Its Regulation by Posttranslational Modification.
Structure, 16, 2008
|
|
1MG5
 
 | | Crystal structure of Drosophila melanogaster alcohol dehydrogenase complexed with NADH and acetate at 1.6 A | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, alcohol dehydrogenase | | Authors: | Benach, J, Atrian, S, Gonzalez-Duarte, R, Ladenstein, R. | | Deposit date: | 2002-08-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Drosophila alcohol dehydrogenase: acetate-enzyme interactions and novel insights into the effects of electrostatics on catalysis
J.Mol.Biol., 345, 2005
|
|
2FQ3
 
 | | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes | | Descriptor: | Transcription regulatory protein SWI3 | | Authors: | Da, G, Lenkart, J, Zhao, K, Shiekhattar, R, Cairns, B.R, Marmorstein, R. | | Deposit date: | 2006-01-17 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2IV0
 
 | | Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure analysis and engineering of chimers | | Descriptor: | CHLORIDE ION, ISOCITRATE DEHYDROGENASE, ZINC ION | | Authors: | Stokke, R, Karlstrom, M, Yang, N, Leiros, I, Ladenstein, R, Birkeland, N.K, Steen, I.H. | | Deposit date: | 2006-06-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Thermal Stability of Isocitrate Dehydrogenase from Archaeoglobus Fulgidus Studied by Crystal Structure Analysis and Engineering of Chimers
Extremophiles, 11, 2007
|
|
4C4K
 
 | | Crystal structure of the titin M10-Obscurin Ig domain 1 complex | | Descriptor: | 1,2-ETHANEDIOL, OBSCURIN, TITIN | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2013-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of the Human Titin:Obscurin Complex Reveals a Conserved Yet Specific Muscle M-Band Zipper Module.
J.Mol.Biol., 427, 2015
|
|
1PKV
 
 | | The N-terminal domain of riboflavin synthase in complex with riboflavin | | Descriptor: | RIBOFLAVIN, Riboflavin synthase alpha chain | | Authors: | Meining, W, Eberhardt, S, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the N-terminal domain of riboflavin synthase in complex with riboflavin at 2.6A resolution.
J.Mol.Biol., 331, 2003
|
|
2JFB
 
 | | 3D Structure of Lumazine Synthase from Candida albicans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, PHOSPHATE ION | | Authors: | Morgunova, E, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lumazine Synthase from Candida Albicans as an Anti- Fungal Target Enzyme: Structural and Biochemical Basis for Drug Design.
J.Biol.Chem., 282, 2007
|
|
3KSL
 
 | | Structure of FPT bound to DATFP-DH-GPP | | Descriptor: | (2S,6E)-8-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}-2,6-dimethyloct-6-en-1-yl (2S)-3,3,3-trifluoro-2-hydrazinopropanoate, Farnesyltransferase, CAAX box, ... | | Authors: | Hovlid, M.L, Edelstein, R.L, Henry, O, Ochocki, J, DeGraw, A, Lenevich, S, Talbot, T, Young, V, Hruza, A.W, Lopez-Gallego, F, Labello, N.P, Strickland, C.L, Schmidt-Dannert, C, Distefano, M.D. | | Deposit date: | 2009-11-23 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis, properties, and applications of diazotrifluropropanoyl-containing photoactive analogs of farnesyl diphosphate containing modified linkages for enhanced stability.
Chem.Biol.Drug Des., 75, 2010
|
|
1SZD
 
 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4F2U
 
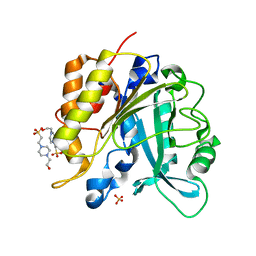 | | Structure of the N254Y/H258Y double mutant of the Phosphatidylinositol-Specific Phospholipase C from S.aureus | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION | | Authors: | Cheng, J, Goldstein, R, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Competition between Anion Binding and Dimerization Modulates Staphylococcus aureus Phosphatidylinositol-specific Phospholipase C Enzymatic Activity.
J.Biol.Chem., 287, 2012
|
|
4F2T
 
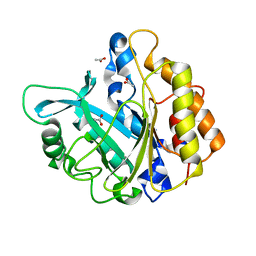 | | Modulation of S.aureus Phosphatidylinositol-Specific Phospholipase C Membrane Binding. | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | Cheng, J, Goldstein, R, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Competition between Anion Binding and Dimerization Modulates Staphylococcus aureus Phosphatidylinositol-specific Phospholipase C Enzymatic Activity.
J.Biol.Chem., 287, 2012
|
|
1QEY
 
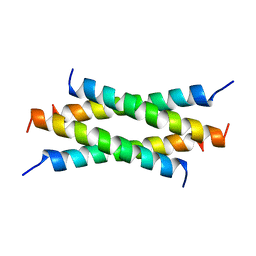 | | NMR Structure Determination of the Tetramerization Domain of the MNT Repressor: An Asymmetric A-Helical Assembly in Slow Exchange | | Descriptor: | PROTEIN (REGULATORY PROTEIN MNT) | | Authors: | Nooren, I.M.A, George, A.V.E, Kaptein, R, Sauer, R.T, Boelens, R. | | Deposit date: | 1999-04-03 | | Release date: | 1999-08-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The tetramerization domain of the Mnt repressor consists of two right-handed coiled coils.
Nat.Struct.Biol., 6, 1999
|
|
3R6B
 
 | | Crystal Structure of Thrombospondin-1 TSR Domains 2 and 3 | | Descriptor: | 1,2-ETHANEDIOL, Thrombospondin-1 | | Authors: | Page, R.C, Klenotic, P.A, Misra, S, Silverstein, R.L. | | Deposit date: | 2011-03-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expression, purification and structural characterization of functionally replete thrombospondin-1 type 1 repeats in a bacterial expression system.
Protein Expr.Purif., 80, 2011
|
|
1QSR
 
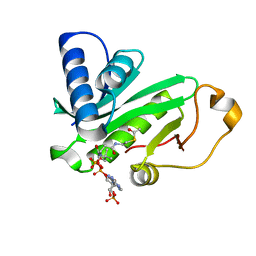 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND ACETYL-COENZYME A | | Descriptor: | ACETYL COENZYME *A, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
