5JT2
 
 | | BRAFV600E Kinase Domain In Complex with Chemically Linked Vemurafenib Inhibitor VEM-BISAMIDE | | Descriptor: | 2,2'-oxybis(N-{[4-(3-{2,6-difluoro-3-[(propane-1-sulfonyl)amino]benzoyl}-1H-pyrrolo[2,3-b]pyridin-5-yl)phenyl]methyl}acetamide), BENZAMIDINE, Serine/threonine-protein kinase B-raf | | Authors: | Grasso, M.J, Marmorstein, R. | | Deposit date: | 2016-05-09 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Chemically Linked Vemurafenib Inhibitors Promote an Inactive BRAF(V600E) Conformation.
Acs Chem.Biol., 11, 2016
|
|
6TTR
 
 | | Crystal Structure of the coiled coil and GGDEF domain of DgcB from Caulobacter crescentus in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GGDEF diguanylate cyclase DgcB, PHOSPHATE ION | | Authors: | Holzschuh, F, Schirmer, T, Teixeira, R.D. | | Deposit date: | 2019-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the coiled coil and GGDEF domain of DgcB from Caulobacter crescentus in complex with c-di-GMP
To Be Published
|
|
6TTS
 
 | | Crystal structure of the GGDEF domain of DgcB from Caulobacter crescentus in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GGDEF diguanylate cyclase DgcB, SULFATE ION | | Authors: | Holzschuh, F, Schirmer, T, Teixeira, R. | | Deposit date: | 2019-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the GGDEF domain of DgcB from Caulobacter crescentus in complex with c-di-GMP
To Be Published
|
|
7RB3
 
 | |
6UIA
 
 | |
8G0L
 
 | |
4PZR
 
 | |
4PZS
 
 | |
4KVO
 
 | |
4KVM
 
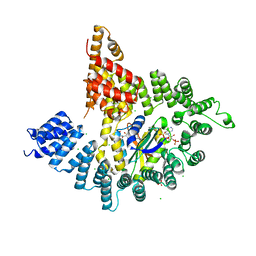 | |
4R5K
 
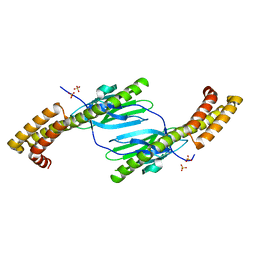 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-B) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, SULFATE ION | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7469 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
5SZR
 
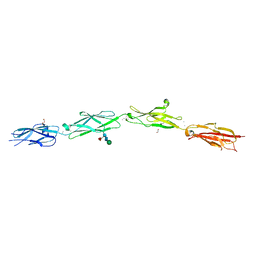 | | Protocadherin Gamma B2 extracellular cadherin domains 3-6 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Rubinstein, R, Honig, B, Shapiro, L. | | Deposit date: | 2016-08-14 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
4R5J
 
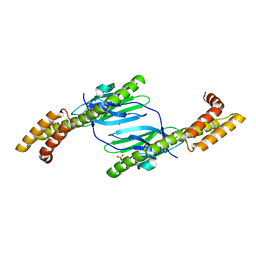 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-A) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, PHOSPHATE ION | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
5FM4
 
 | |
4R5G
 
 | | Crystal structure of the DnaK C-terminus with the inhibitor PET-16 | | Descriptor: | Chaperone protein DnaK, triphenyl(phenylethynyl)phosphonium | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4501 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
4R5I
 
 | | Crystal structure of the DnaK C-terminus with the substrate peptide NRLLLTG | | Descriptor: | Chaperone protein DnaK, HSP70/DnaK Substrate Peptide: NRLLLTG, PHOSPHATE ION, ... | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9702 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
7KD7
 
 | |
7KPU
 
 | |
4ZPO
 
 | | Crystal Structure of Protocadherin Gamma C5 EC1-3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MCG133388, ... | | Authors: | Wolcott, H.N, Goodman, K.M, Bahna, F, Mannepalli, S, Rubinstein, R, Honig, B, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
4KVX
 
 | | Crystal structure of Naa10 (Ard1) bound to AcCoA | | Descriptor: | ACETYL COENZYME *A, N-terminal acetyltransferase A complex catalytic subunit ard1 | | Authors: | Liszczak, G.P, Marmorstein, R.Q. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for N-terminal acetylation by the heterodimeric NatA complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
8RWK
 
 | | cryoEM structure of the central Ald4 filament determined by FilamentID | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium-activated aldehyde dehydrogenase, mitochondrial | | Authors: | Hugener, J, Xu, J, Wettstein, R, Ioannidi, L, Velikov, D, Wollweber, F, Henggeler, A, Matos, J, Pilhofer, M. | | Deposit date: | 2024-02-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | FilamentID reveals the composition and function of metabolic enzyme polymers during gametogenesis.
Cell, 187, 2024
|
|
1XGV
 
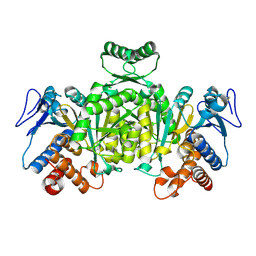 | | Isocitrate Dehydrogenase from the hyperthermophile Aeropyrum pernix | | Descriptor: | Isocitrate dehydrogenase | | Authors: | Karlstrom, M, Stokke, R, Steen, I.H, Birkeland, N.-K, Ladenstein, R. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isocitrate dehydrogenase from the hyperthermophile Aeropyrum pernix: X-ray structure analysis of a ternary enzyme-substrate complex and thermal stability
J.Mol.Biol., 345, 2005
|
|
7LJ9
 
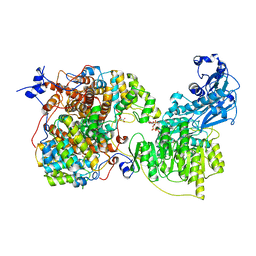 | |
1ZX2
 
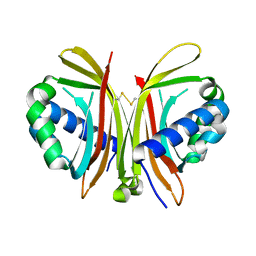 | | Crystal Structure of Yeast UBP3-associated Protein BRE5 | | Descriptor: | UBP3-associated protein BRE5 | | Authors: | Li, K, Zhao, K, Ossareh-Nazari, B, Da, G, Dargemont, C, Marmorstein, R. | | Deposit date: | 2005-06-06 | | Release date: | 2005-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for interaction between the Ubp3 deubiquitinating enzyme and its Bre5 cofactor
J.Biol.Chem., 280, 2005
|
|
3ZFW
 
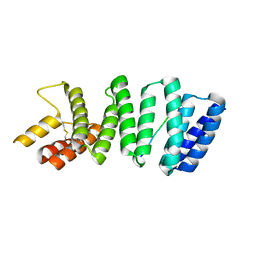 | | Crystal structure of the TPR domain of kinesin light chain 2 in complex with a tryptophan-acidic cargo peptide | | Descriptor: | KINESIN LIGHT CHAIN 2, PLECKSTRIN HOMOLOGY DOMAIN-CONTAINING FAMILY M MEMBER 2 | | Authors: | Pernigo, S, Lamprecht, A, Steiner, R.A, Dodding, M.P. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Kinesin-1:Cargo Recognition.
Science, 340, 2013
|
|
