8B5B
 
 | | Human BRD4 bromdomain 1 in complex with a H4 peptide containing acetyl lysine and ApmTri (H4K5acK8ApmTri) | | Descriptor: | Bromodomain-containing protein 4, DI(HYDROXYETHYL)ETHER, H4K5acK8ApmTri | | Authors: | Braun, M.B, Bartlick, N, Stehle, T. | | Deposit date: | 2022-09-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Synthesis, Biochemical Characterization, and Genetic Encoding of a 1,2,4-Triazole Amino Acid as an Acetyllysine Mimic for Bromodomains of the BET Family.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8B5A
 
 | | Human BRD3 bromodomain 2 in complex with a H4 peptide containing ApmTri (H4K20ApmTri) | | Descriptor: | Bromodomain-containing protein 3, H4K20ApmTri | | Authors: | Braun, M.B, Bartlick, N, Stehle, T. | | Deposit date: | 2022-09-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Synthesis, Biochemical Characterization, and Genetic Encoding of a 1,2,4-Triazole Amino Acid as an Acetyllysine Mimic for Bromodomains of the BET Family.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8B5C
 
 | | Human BRD4 bromdomain 1 in complex with a H4 peptide containing ApmTri (H4K5/8ApmTri) | | Descriptor: | Bromodomain-containing protein 4, H4K5/8ApmTri | | Authors: | Braun, M.B, Bartlick, N, Stehle, T. | | Deposit date: | 2022-09-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Synthesis, Biochemical Characterization, and Genetic Encoding of a 1,2,4-Triazole Amino Acid as an Acetyllysine Mimic for Bromodomains of the BET Family.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4ZH1
 
 | | Complement factor H in complex with the GM1 glycan | | Descriptor: | Complement C3, Complement factor H, GLYCEROL, ... | | Authors: | Blaum, B.S, Stehle, T. | | Deposit date: | 2015-04-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Complement Factor H and Simian Virus 40 bind the GM1 ganglioside in distinct conformations.
Glycobiology, 26, 2016
|
|
4PCG
 
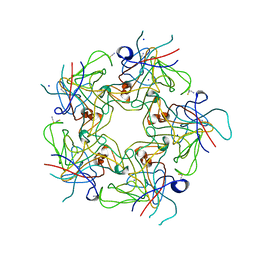 | | Structure of Human Polyomavirus 6 (HPyV6) VP1 pentamer | | Descriptor: | SODIUM ION, THIOCYANATE ION, VP1 | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-04-15 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure analysis of the major capsid proteins of human polyomaviruses 6 and 7 reveals an obstructed sialic Acid binding site.
J.Virol., 88, 2014
|
|
4ZXZ
 
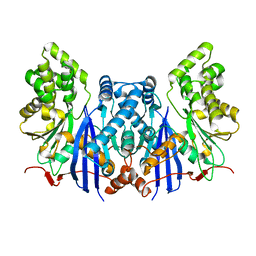 | |
4FMJ
 
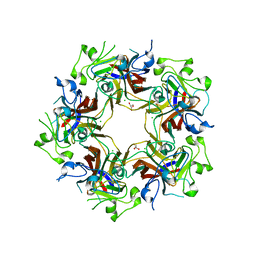 | | Merkel cell polyomavirus VP1 in complex with GD1a oligosaccharide | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Neu, U, Hengel, H, Stehle, T. | | Deposit date: | 2012-06-17 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Merkel Cell Polyomavirus VP1 Complexes Define a Sialic Acid Binding Site Required for Infection.
Plos Pathog., 8, 2012
|
|
1M1X
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN ALPHA VBETA3 BOUND TO MN2+ | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.-P, Stehle, T, Zhang, R, Joachimiak, A, Frech, M, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand.
Science, 296, 2002
|
|
3MK4
 
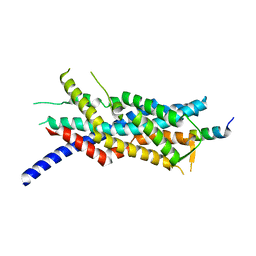 | | X-Ray structure of human PEX3 in complex with a PEX19 derived peptide | | Descriptor: | Peroxisomal biogenesis factor 19, Peroxisomal biogenesis factor 3 | | Authors: | Schmidt, F, Treiber, N, Dodt, G, Stehle, T. | | Deposit date: | 2010-04-14 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Insights into peroxisome function from the structure of PEX3 in complex with a soluble fragment of PEX19
J.Biol.Chem., 285, 2010
|
|
2O39
 
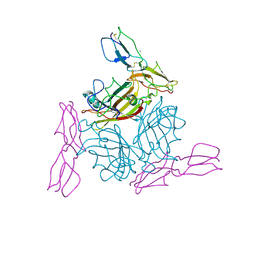 | | Human Adenovirus type 11 knob in complex with domains SCR1 and SCR2 of CD46 (membrane cofactor protein, MCP) | | Descriptor: | CALCIUM ION, Fiber protein, Membrane cofactor protein, ... | | Authors: | Persson, D.B, Reiter, D.M, Arnberg, N, Stehle, T. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Adenovirus type 11 binding alters the conformation of its receptor CD46.
Nat.Struct.Mol.Biol., 14, 2007
|
|
3O24
 
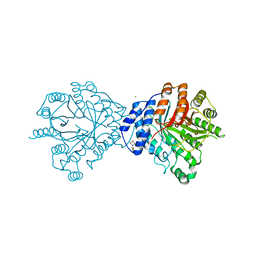 | | Crystal structure of the brevianamide F prenyltransferase FtmPT1 from Aspergillus fumigatus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Brevianamide F prenyltransferase, CHLORIDE ION, ... | | Authors: | Jost, M, Zocher, G.E, Stehle, T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function analysis of an enzymatic prenyl transfer reaction identifies a reaction chamber with modifiable specificity.
J.Am.Chem.Soc., 132, 2010
|
|
4Q4Y
 
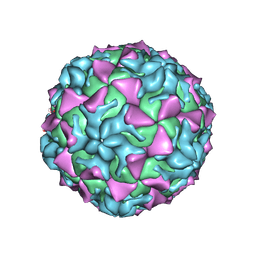 | |
4Q4X
 
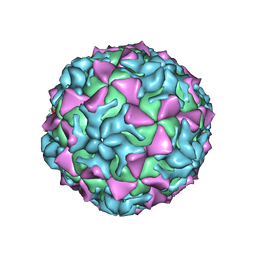 | |
1M1U
 
 | | AN ISOLEUCINE-BASED ALLOSTERIC SWITCH CONTROLS AFFINITY AND SHAPE SHIFTING IN INTEGRIN CD11B A-DOMAIN | | Descriptor: | CALCIUM ION, Integrin alpha-M | | Authors: | Xiong, J.-P, Li, R, Essafi, M, Stehle, T, Arnaout, M.A. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An isoleucine-based allosteric switch controls affinity and shape shifting in integrin CD11b A-domain.
J.Biol.Chem., 275, 2000
|
|
3O2K
 
 | | Crystal Structure of Brevianamide F Prenyltransferase Complexed with Brevianamide F and Dimethylallyl S-thiolodiphosphate | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Brevianamide F prenyltransferase, ... | | Authors: | Jost, M, Zocher, G.E, Stehle, T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-function analysis of an enzymatic prenyl transfer reaction identifies a reaction chamber with modifiable specificity.
J.Am.Chem.Soc., 132, 2010
|
|
3T6S
 
 | | Crystal structure of CerJ from Streptomyces tendae in Complex with CoA | | Descriptor: | COENZYME A, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-07-29 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
5CPU
 
 | | Crystal structure of murine polyomavirus PTA strain VP1 | | Descriptor: | GLYCEROL, VP1 | | Authors: | Liaci, A.M, Buch, M.H.C, Neu, U, Stehle, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.
Plos Pathog., 11, 2015
|
|
5CPX
 
 | | Crystal structure of murine polyomavirus PTA strain VP1 in complex with the DSLNT glycan | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Buch, M.H.C, Liaci, A.M, Neu, U, Stehle, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.
Plos Pathog., 11, 2015
|
|
3T8E
 
 | | Crystal structure of CerJ from Streptomyces tendae soaked with CerviK | | Descriptor: | ACETATE ION, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-08-01 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
3T5Y
 
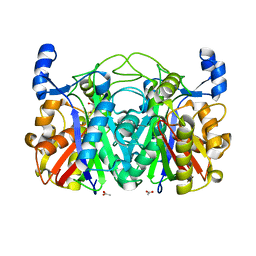 | | Crystal structure of CerJ from Streptomyces tendae - malonic acid covalently linked to the catalytic Cystein C116 | | Descriptor: | ACETATE ION, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
5EMI
 
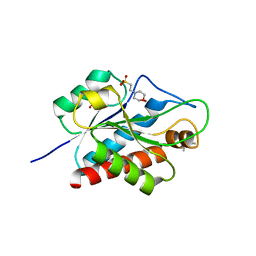 | | N-acetylmuramoyl-L-alanine amidase AmiC2 of Nostoc punctiforme | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cell wall hydrolase/autolysin, ... | | Authors: | Buettner, F.M, Stehle, T. | | Deposit date: | 2015-11-06 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Enabling cell-cell communication via nanopore formation: structure, function and localization of the unique cell wall amidase AmiC2 of Nostoc punctiforme.
Febs J., 283, 2016
|
|
4MBZ
 
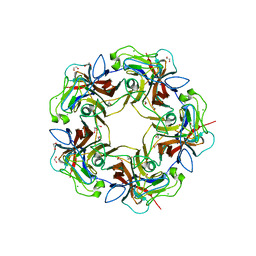 | | Structure of B-Lymphotropic Polyomavirus VP1 in complex with 3'-sialyllactosamine | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Khan, Z.M, Neu, U, Stehle, T. | | Deposit date: | 2013-08-21 | | Release date: | 2013-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of B-Lymphotropic Polyomavirus VP1 in Complex with Oligosaccharide Ligands.
Plos Pathog., 9, 2013
|
|
4MBX
 
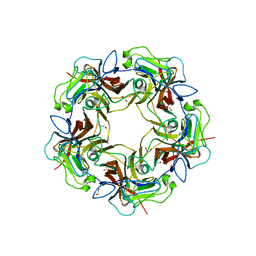 | | Structure of unliganded B-Lymphotropic Polyomavirus VP1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Major Capsid Protein VP1 | | Authors: | Khan, Z.M, Neu, U, Schuch, B, Stehle, T. | | Deposit date: | 2013-08-21 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of B-Lymphotropic Polyomavirus VP1 in Complex with Oligosaccharide Ligands.
Plos Pathog., 9, 2013
|
|
3S7X
 
 | | Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1, ... | | Authors: | Neu, U, Wang, J, Stehle, T. | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the major capsid proteins of the human KI and WU polyomaviruses.
J.Virol., 85, 2011
|
|
3S7V
 
 | | Unassembled KI Polyomavirus VP1 Pentamer | | Descriptor: | Major capsid protein VP1 | | Authors: | Neu, U, Stehle, T. | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of the major capsid proteins of the human KI and WU polyomaviruses.
J.Virol., 85, 2011
|
|
