2VCT
 
 | |
2WJU
 
 | |
8UW1
 
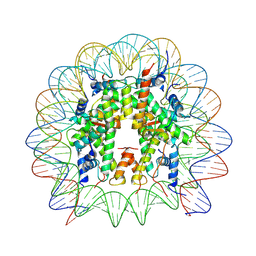 | | Cryo-EM structure of DNMT3A1 UDR in complex with H2AK119Ub-nucleosome | | Descriptor: | DNA (146-MER), DNA (cytosine-5)-methyltransferase 3A, Histone H2A, ... | | Authors: | Gretarsson, K, Abini-Agbomson, S, Armache, K.-J, Lu, C. | | Deposit date: | 2023-11-05 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cancer-associated DNA hypermethylation of Polycomb targets requires DNMT3A dual recognition of histone H2AK119 ubiquitination and the nucleosome acidic patch.
Sci Adv, 10, 2024
|
|
6Z2T
 
 | |
8CQO
 
 | | Crystal structure of Borrelia burgdorferi paralogous family 12 outer surface protein BBK01 (Se-Met data) | | Descriptor: | Lipoprotein, putative | | Authors: | Brangulis, K, Drunka, L, Matisone, S, Akopjana, I, Tars, K. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Members of the paralogous gene family 12 from the Lyme disease agent Borrelia burgdorferi are non-specific DNA-binding proteins.
Plos One, 19, 2024
|
|
8CQN
 
 | | Crystal structure of Borrelia burgdorferi paralogous family 12 outer surface protein BBK01 | | Descriptor: | Lipoprotein, putative | | Authors: | Brangulis, K, Drunka, L, Matisone, S, Akopjana, I, Tars, K. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Members of the paralogous gene family 12 from the Lyme disease agent Borrelia burgdorferi are non-specific DNA-binding proteins.
Plos One, 19, 2024
|
|
1X27
 
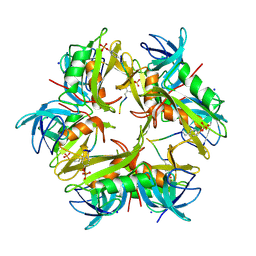 | | Crystal Structure of Lck SH2-SH3 with SH2 binding site of p130Cas | | Descriptor: | CRK-associated substrate, Proto-oncogene tyrosine-protein kinase LCK, SODIUM ION | | Authors: | Nasertorabi, F, Tars, K, Becherer, K, Kodandapani, R, Liljas, L, Vuori, K, Ely, K.R. | | Deposit date: | 2005-04-20 | | Release date: | 2006-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for regulation of Src by the docking protein p130Cas
J.MOL.RECOG., 19, 2006
|
|
5BWY
 
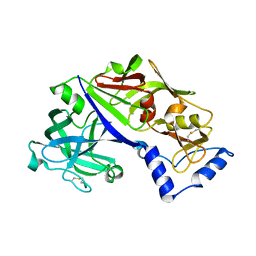 | | Structure of proplasmepsin II from Plasmodium falciparum, Space Group P43212 | | Descriptor: | Plasmepsin-2 | | Authors: | Recacha, R, Akopjana, I, Tars, K, Jaudzems, K. | | Deposit date: | 2015-06-08 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Crystal structure of Plasmodium falciparum proplasmepsin IV: the plasticity of proplasmepsins.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
6HPN
 
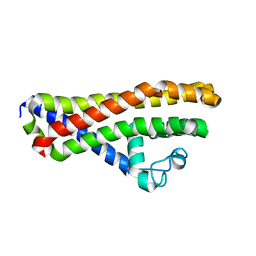 | | Crystal structure of Borrelia spielmanii WP_012665240 (BSA64) - an orthologous protein to B. burgdorferi BBA64 | | Descriptor: | Antigen, P35 | | Authors: | Brangulis, K, Akopjana, I, Kazaks, A, Tars, K. | | Deposit date: | 2018-09-21 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the outer surface proteins from Borrelia burgdorferi paralogous gene family 54 that are thought to be the key players in the pathogenesis of Lyme disease.
J.Struct.Biol., 210, 2020
|
|
4L8H
 
 | |
5JOD
 
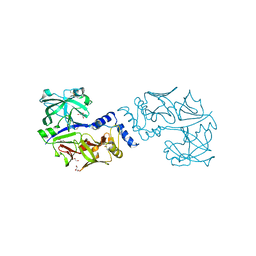 | | Structure of proplasmepsin IV from Plasmodium falciparum | | Descriptor: | GLYCEROL, Proplasmepsin IV | | Authors: | Recacha, R, Akopjana, I, Tars, K, Jaudzems, K. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Crystal structure of Plasmodium falciparum proplasmepsin IV: the plasticity of proplasmepsins.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
8CO0
 
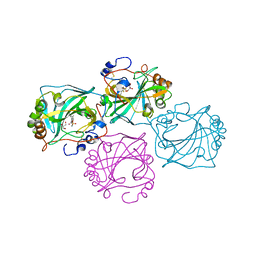 | |
5AMD
 
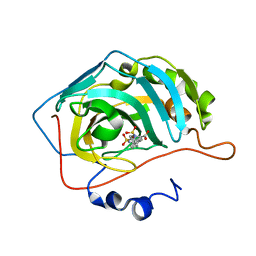 | |
1FR5
 
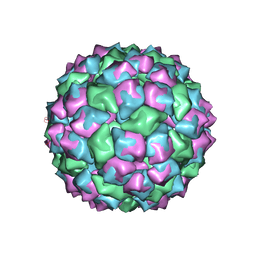 | | PHAGE FR CAPSIDS WITH A FOUR RESIDUE DELETION IN THE COAT PROTEIN FG LOOP | | Descriptor: | BACTERIOPHAGE FR CAPSID | | Authors: | Axblom, C, Tars, K, Fridborg, K, Bundule, M, Orna, L, Liljas, L. | | Deposit date: | 1998-07-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of phage fr capsids with a deletion in the FG loop: implications for viral assembly.
Virology, 249, 1998
|
|
5LQP
 
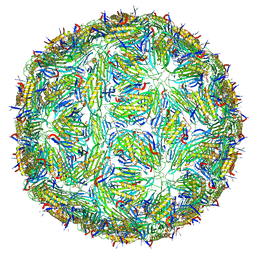 | |
5JXV
 
 | | Solid-state MAS NMR structure of immunoglobulin beta 1 binding domain of protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Andreas, L.B, Jaudzems, K, Stanek, J, Lalli, D, Bertarello, A, Le Marchand, T, Cala-De Paepe, D, Kotelovica, S, Akopjana, I, Knott, B, Wegner, S, Engelke, F, Lesage, A, Emsley, L, Tars, K, Herrmann, T, Pintacuda, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure of fully protonated proteins by proton-detected magic-angle spinning NMR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JZR
 
 | | Solid-state MAS NMR structure of Acinetobacter phage 205 (AP205) coat protein in assembled capsid particles | | Descriptor: | Coat protein | | Authors: | Jaudzems, K, Andreas, L.B, Stanek, J, Lalli, D, Bertarello, A, Le Marchand, T, Cala-De Paepe, D, Kotelovica, S, Akopjana, I, Knott, B, Wegner, S, Engelke, F, Lesage, A, Emsley, L, Tars, K, Herrmann, T, Pintacuda, G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure of fully protonated proteins by proton-detected magic-angle spinning NMR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3N6W
 
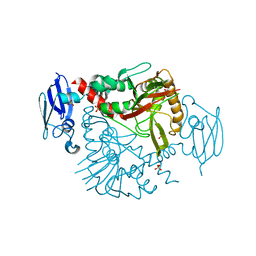 | | Crystal structure of human gamma-butyrobetaine hydroxylase | | Descriptor: | Gamma-butyrobetaine dioxygenase, SULFATE ION, ZINC ION | | Authors: | Rumnieks, J, Zeltins, A, Leonchiks, A, Kazaks, A, Kotelovica, S, Tars, K. | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human gamma-butyrobetaine hydroxylase.
Biochem.Biophys.Res.Commun., 398, 2010
|
|
1ZSE
 
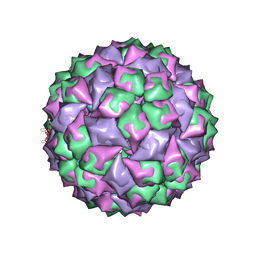 | | RNA stemloop from bacteriophage Qbeta complexed with an N87S mutant MS2 Capsid | | Descriptor: | Coat protein, RNA HAIRPIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-24 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
5FS4
 
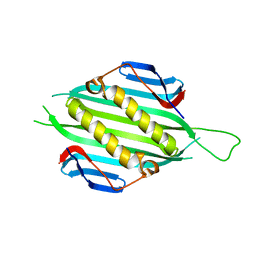 | | Bacteriophage AP205 coat protein | | Descriptor: | AP205 BACTERIOPHAGE COAT PROTEIN | | Authors: | Shishovs, M, Tars, K. | | Deposit date: | 2015-12-29 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of Ap205 Coat Protein Reveals Circular Permutation in Ssrna Bacteriophages.
J.Mol.Biol., 428, 2016
|
|
5FL6
 
 | | Three dimensional structure of human carbonic anhydrase IX in complex with 5-(1-(4-Methylphenyl)-1H-1,2,3-triazol-4-yl)thiophene-2- sulfonamide | | Descriptor: | 5-[1-(4-methylphenyl)-1,2,3-triazol-4-yl]thiophene-2-sulfonamide, ACETIC ACID, CARBONIC ANHYDRASE IX, ... | | Authors: | Leitans, J, Tars, K, Zalubovskis, R. | | Deposit date: | 2015-10-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An Efficient Expression and Crystallization System of the Cancer Asociated Carbonic Anhydrase Isoform Ix.
J.Med.Chem., 58, 2015
|
|
5FL5
 
 | | Three dimensional structure of human carbonic anhydrase IX in complex with 5-(1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)thiophene-2- sulfonamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[1-(4-methoxyphenyl)-1,2,3-triazol-4-yl]thiophene-2-sulfonamide, CARBONIC ANHYDRASE IX, ... | | Authors: | Leitans, J, Tars, K, Zalubovskis, R. | | Deposit date: | 2015-10-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An Efficient Expression and Crystallization System of the Cancer Asociated Carbonic Anhydrase Isoform Ix.
J.Med.Chem., 58, 2015
|
|
5FL4
 
 | | Three dimensional structure of human carbonic anhydrase IX in complex with 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide | | Descriptor: | 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide, ACETIC ACID, CARBONIC ANHYDRASE 9, ... | | Authors: | Leitans, J, Tars, K, Zalubovskis, R. | | Deposit date: | 2015-10-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | An Efficient Expression and Crystallization System of the Cancer Asociated Carbonic Anhydrase Isoform Ix.
J.Med.Chem., 58, 2015
|
|
3RLC
 
 | |
3RLK
 
 | |
