3LQB
 
 | | Crystal structure of the hatching enzyme ZHE1 from the zebrafish Danio rerio | | Descriptor: | 1,2-ETHANEDIOL, LOC792177 protein, SULFATE ION, ... | | Authors: | Tanokura, M, Okada, A, Nagata, K, Yasumasu, S, Ohtsuka, J, Iuchi, I. | | Deposit date: | 2010-02-08 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of zebrafish hatching enzyme 1 from the zebrafish Danio rerio
J.Mol.Biol., 402, 2010
|
|
2E3U
 
 | |
3AEV
 
 | | Crystal structure of a/eIF2alpha-aDim2p-rRNA complex from Pyrococcus horikoshii OT3 | | Descriptor: | Putative uncharacterized protein PH1566, RNA (5'-R(*GP*GP*AP*UP*CP*AP*CP*CP*UP*CP*C)-3'), Translation initiation factor 2 subunit alpha | | Authors: | Tanokura, M, Jia, M.Z, Nagata, K. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An archaeal Dim2-like protein, aDim2p, forms a ternary complex with a/eIF2 alpha and the 3' end fragment of 16S rRNA
J.Mol.Biol., 398, 2010
|
|
9IHS
 
 | | Microbial transglutaminase mutant - D3C/G283C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Suzuki, M, Date, M, Kashiwagi, T, Takahashi, K, Nakamura, A, Tanokura, M, Suzuki, E, Yokoyama, K. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Random mutagenesis and disulfide bond formation improved thermostability in microbial transglutaminase.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
5WVU
 
 | | Crystal structure of carboxypeptidase from Thermus thermophilus | | Descriptor: | GLYCEROL, Thermostable carboxypeptidase 1, ZINC ION | | Authors: | Okai, M, Nagata, K, Tanokura, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the transition between the open and closed conformations of Thermus thermophilus carboxypeptidase.
Biochem. Biophys. Res. Commun., 484, 2017
|
|
5WUT
 
 | |
6L2N
 
 | | Crystal structure of the R.PabI(Y68F-K154A)-dsDNA(GTAC-3bp-GTAC) complex | | Descriptor: | DNA (5'-D(*TP*CP*AP*GP*CP*AP*GP*TP*AP*CP*TP*AP*AP*GP*TP*AP*CP*TP*GP*CP*TP*GP*A)-3'), RE_R_Pab1 domain-containing protein | | Authors: | Miyazono, K, Wang, D, Ito, T, Tanokura, M. | | Deposit date: | 2019-10-05 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI.
Nucleic Acids Res., 48, 2020
|
|
6L2O
 
 | | Crystal structure of the R.PabI(Y68F-K154A)-dsDNA(GTAC-5bp-GTAC) complex | | Descriptor: | DNA (5'-D(*CP*A*GP*CP*AP*GP*TP*AP*CP*TP*TP*AP*AP*AP*GP*TP*AP*CP*TP*GP*CP*TP*G)-3'), RE_R_Pab1 domain-containing protein | | Authors: | Miyazono, K, Wang, D, Ito, T, Tanokura, M. | | Deposit date: | 2019-10-05 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI.
Nucleic Acids Res., 48, 2020
|
|
5TLC
 
 | |
2RR7
 
 | | Microtubule Binding Domain of DYNEIN-C | | Descriptor: | Dynein heavy chain 9 | | Authors: | Kato, Y, Yagi, T, Ohki, S, Burgess, S, Honda, S, Kamiya, R, Tanokura, M. | | Deposit date: | 2010-06-04 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the microtubule-binding domain of flagellar dynein
Structure, 22, 2014
|
|
7BQU
 
 | | Cereblon in complex with SALL4 and (S)-thalidomide | | Descriptor: | Protein cereblon, S-Thalidomide, Sal-like protein 4, ... | | Authors: | Furihata, H, Miyauchi, Y, Asano, A, Tanokura, M, Miyakawa, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural bases of IMiD selectivity that emerges by 5-hydroxythalidomide.
Nat Commun, 11, 2020
|
|
7BQV
 
 | | Cereblon in complex with SALL4 and (S)-5-hydroxythalidomide | | Descriptor: | 2-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-5-oxidanyl-isoindole-1,3-dione, Protein cereblon, SULFATE ION, ... | | Authors: | Furihata, H, Miyauchi, Y, Asano, A, Tanokura, M, Miyakawa, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural bases of IMiD selectivity that emerges by 5-hydroxythalidomide.
Nat Commun, 11, 2020
|
|
3JRS
 
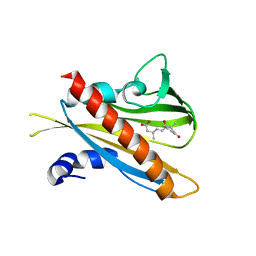 | | Crystal structure of (+)-ABA-bound PYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
3JRQ
 
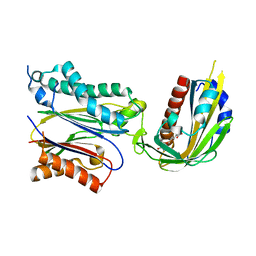 | | Crystal structure of (+)-ABA-bound PYL1 in complex with ABI1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Protein phosphatase 2C 56, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
4IJ5
 
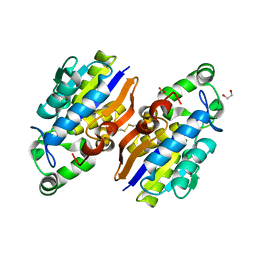 | | Crystal Structure of a Novel-type Phosphoserine Phosphatase from Hydrogenobacter thermophilus TK-6 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Phosphoserine phosphatase 1 | | Authors: | Chiba, Y, Horita, S, Ohtsuka, J, Arai, H, Nagata, K, Igarashi, Y, Tanokura, M, Ishii, M. | | Deposit date: | 2012-12-21 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural units important for activity of a novel-type phosphoserine phosphatase from Hydrogenobacter thermophilus TK-6 revealed by crystal structure analysis
J.Biol.Chem., 288, 2013
|
|
4IJ6
 
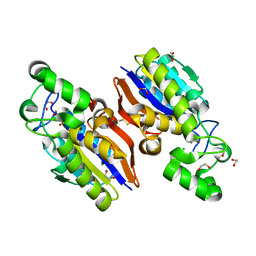 | | Crystal Structure of a Novel-type Phosphoserine Phosphatase Mutant (H9A) from Hydrogenobacter thermophilus TK-6 in Complex with L-phosphoserine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHOSERINE, ... | | Authors: | Chiba, Y, Horita, S, Ohtsuka, J, Arai, H, Nagata, K, Igarashi, Y, Tanokura, M, Ishii, M. | | Deposit date: | 2012-12-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural units important for activity of a novel-type phosphoserine phosphatase from Hydrogenobacter thermophilus TK-6 revealed by crystal structure analysis
J.Biol.Chem., 288, 2013
|
|
6M64
 
 | | Crystal structure of SMAD2 in complex with CBP | | Descriptor: | CBP, Mothers against decapentaplegic homolog 2 | | Authors: | Miyazono, K, Ito, T, Wada, H, Tanokura, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for transcriptional coactivator recognition by SMAD2 in TGF-beta signaling.
Sci.Signal., 13, 2020
|
|
6M3L
 
 | | Crystal structure of the R.PabI(Y68F-K154A)-dsDNA(nonspecific) complex | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*CP*GP*AP*TP*TP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*GP*CP*G)-3'), RE_R_Pab1 domain-containing protein | | Authors: | Miyazono, K, Wang, D, Ito, T, Tanokura, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI.
Nucleic Acids Res., 48, 2020
|
|
6M6P
 
 | | Structure of Marine bacterial laminarinase mutant E135A in complex with 1,3-beta-cellotriosyl-glucose | | Descriptor: | CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose, laminarinase | | Authors: | Yang, J, Xu, Y, Tanokura, M, Long, L, Miyakawa, T. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
4WFJ
 
 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-bound state at 1.75 angstrom resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cutinase | | Authors: | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4WFK
 
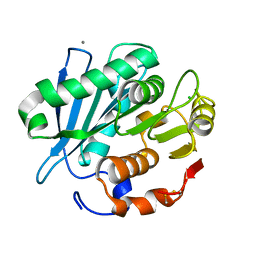 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-bound state at 2.35 angstrom resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cutinase | | Authors: | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4TMC
 
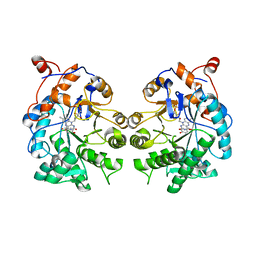 | | CRYSTAL STRUCTURE of OLD YELLOW ENZYME from CANDIDA MACEDONIENSIS AKU4588 COMPLEXED with P-HYDROXYBENZALDEHYDE | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old yellow enzyme, P-HYDROXYBENZALDEHYDE | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2014-05-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Engineered Old Yellow Enzyme that Enables Efficient Synthesis of (4R,6R)-Actinol in a One-Pot Reduction System
Chembiochem, 16, 2015
|
|
4WFI
 
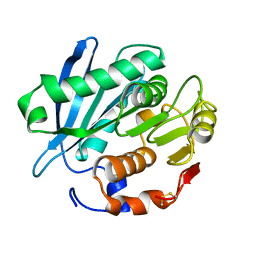 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-free state | | Descriptor: | Cutinase | | Authors: | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4TMB
 
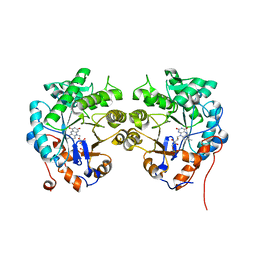 | | CRYSTAL STRUCTURE of OLD YELLOW ENZYME from CANDIDA MACEDONIENSIS AKU4588 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old yellow enzyme | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2014-05-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Engineered Old Yellow Enzyme that Enables Efficient Synthesis of (4R,6R)-Actinol in a One-Pot Reduction System
Chembiochem, 16, 2015
|
|
2GXG
 
 | |
