8DKQ
 
 | |
8DKO
 
 | |
8DKP
 
 | |
6U2X
 
 | | Structure of ALDH7A1 mutant E399G complexed with NAD | | Descriptor: | Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-08-20 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
7SQN
 
 | |
2BKJ
 
 | |
6O4D
 
 | | Structure of ALDH7A1 mutant W175A complexed with L-pipecolic acid | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4B
 
 | | Structure of ALDH7A1 mutant W175G complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4L
 
 | | Structure of ALDH7A1 mutant E399D complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
6O4K
 
 | | Structure of ALDH7A1 mutant E399Q complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
6O4I
 
 | | Structure of ALDH7A1 mutant E399D complexed with alpha-aminoadipate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINOHEXANEDIOIC ACID, Alpha-aminoadipic semialdehyde dehydrogenase | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
7US3
 
 | | Structure of Putrescine N-hydroxylase Involved Complexed with NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putrescine N-hydroxylase, ... | | Authors: | Tanner, J.J, Bogner, A.N. | | Deposit date: | 2022-04-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N -Hydroxylase from Acinetobacter baumannii.
Biochemistry, 61, 2022
|
|
1I8M
 
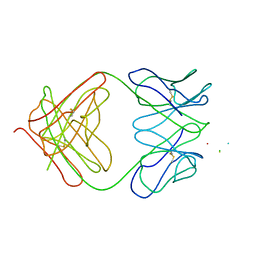 | | CRYSTAL STRUCTURE OF A RECOMBINANT ANTI-SINGLE-STRANDED DNA ANTIBODY FRAGMENT COMPLEXED WITH DT5 | | Descriptor: | 5'-D(*TP*TP*TP*TP*T)-3', 5'-D(P*TP*T)-3', ANTIBODY HEAVY CHAIN FAB, ... | | Authors: | Tanner, J.J, Komissarov, A.A, Deutscher, S.L. | | Deposit date: | 2001-03-14 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of an Antigen-Binding Fragment Bound to Single-Stranded DNA
J.Mol.Biol., 314, 2001
|
|
5JJG
 
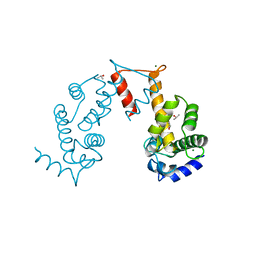 | | Structure of magnesium-loaded ALG-2 | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, Pcalcium-binding protein ALG-2, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2016-04-23 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | EF5 Is the High-Affinity Mg(2+) Site in ALG-2.
Biochemistry, 55, 2016
|
|
5KF6
 
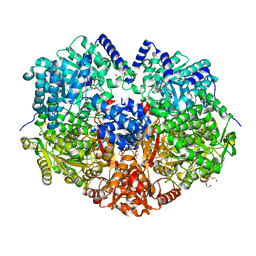 | |
5KOW
 
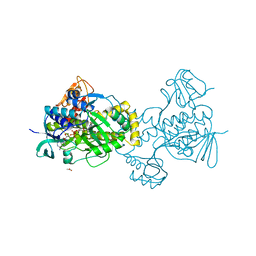 | | Structure of rifampicin monooxygenase | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Pentachlorophenol 4-monooxygenase | | Authors: | Tanner, J.J, Liu, L.-K. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Antibiotic Deactivating, N-hydroxylating Rifampicin Monooxygenase.
J.Biol.Chem., 291, 2016
|
|
5KOX
 
 | |
5KF7
 
 | |
1CER
 
 | |
3ITG
 
 | |
3HAZ
 
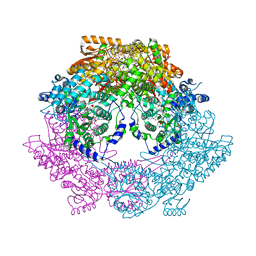 | | Crystal structure of bifunctional proline utilization A (PutA) protein | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2009-05-03 | | Release date: | 2010-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5HHF
 
 | |
7JVK
 
 | |
7JVL
 
 | |
7LRN
 
 | |
