1SBW
 
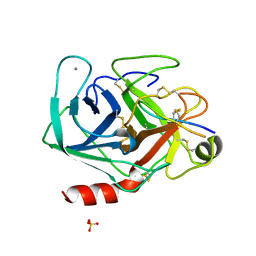 | | CRYSTAL STRUCTURE OF MUNG BEAN INHIBITOR LYSINE ACTIVE FRAGMENT COMPLEX WITH BOVINE BETA-TRYPSIN AT 1.8A RESOLUTION | | Descriptor: | CALCIUM ION, PROTEIN (BETA-TRYPSIN), PROTEIN (MUNG BEAN INHIBITOR LYSIN ACTIVE FRAGMENT), ... | | Authors: | Huang, Q, Zhu, Y, Chi, C, Tang, Y. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of mung bean inhibitor lysine active fragment complex with bovine beta-trypsin at 1.8A resolution.
J.Biomol.Struct.Dyn., 16, 1999
|
|
2LFC
 
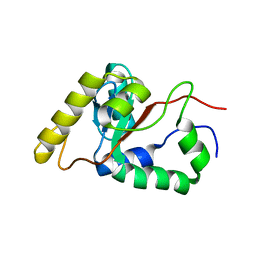 | | Solution NMR Structure of Fumarate reductase flavoprotein subunit from Lactobacillus plantarum, Northeast Structural Genomics Consortium Target LpR145J | | Descriptor: | Fumarate reductase, flavoprotein subunit | | Authors: | Liu, G, Tang, Y, Xiao, R, Janjua, H, Ciccosanti, C, Wang, H, Acton, T, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target LpR145J
To be Published
|
|
6IX8
 
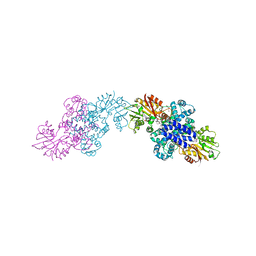 | | The structure of LepI C52A in complex with SAM and its substrate analogue | | Descriptor: | (1R,2R,4aS,8S,8aR)-2,8-dimethyl-5'-phenyl-4a,5,6,7,8,8a-hexahydro-2H,2'H-spiro[naphthalene-1,3'-pyridine]-2',4'(1'H)-dione, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | Deposit date: | 2018-12-09 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
6IX5
 
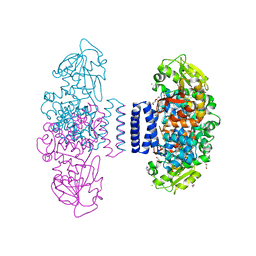 | | The structure of LepI complex with SAM and its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-3-[(2S,6E,8E)-2-methyldeca-6,8-dienoyl]-5-phenylpyridin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | Deposit date: | 2018-12-09 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
6IX3
 
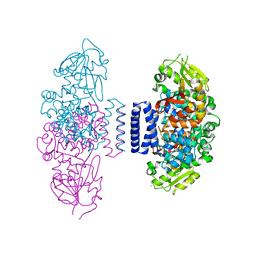 | | The structure of LepI complex with SAM | | Descriptor: | CHLORIDE ION, O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | Authors: | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | Deposit date: | 2018-12-09 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
6IX9
 
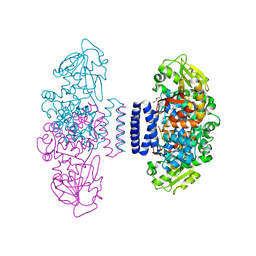 | | The structure of LepI C52A in complex with SAM and leporin C | | Descriptor: | (6R,6aS,10S,10aR)-10-methyl-4-phenyl-6-[(1E)-prop-1-en-1-yl]-2,6,6a,7,8,9,10,10a-octahydro-1H-[2]benzopyrano[4,3-c]pyridin-1-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | Deposit date: | 2018-12-09 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
6IX7
 
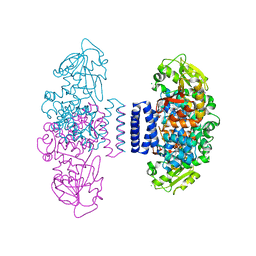 | | The structure of LepI C52A in complex with SAH and substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-3-[(2S,6E,8E)-2-methyldeca-6,8-dienoyl]-5-phenylpyridin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | Deposit date: | 2018-12-09 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
3TL1
 
 | | Crystal structure of the Streptomyces coelicolor WhiE ORFVI polyketide aromatase/cyclase | | Descriptor: | 6,7,9-trihydroxy-3-methyl-1H-benzo[g]isochromen-1-one, GLYCEROL, Polyketide cyclase | | Authors: | Lee, M.-Y, Ames, B.D, Zhang, W, Tang, Y, Tsai, S.-C. | | Deposit date: | 2011-08-29 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insight into the Molecular Basis of Aromatic Polyketide Cyclization: Crystal Structure and in Vitro Characterization of WhiE-ORFVI.
Biochemistry, 51, 2012
|
|
5XJ9
 
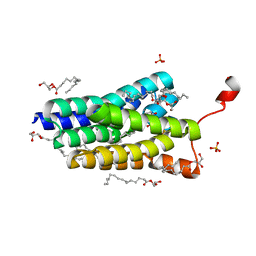 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the orthophosphate form | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION | | Authors: | Li, Z, Tang, Y, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
8HS0
 
 | | The mutant structure of DHAD V178W | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2022-12-16 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The mutant structure of DHAD
To Be Published
|
|
8IMU
 
 | | Dihydroxyacid dehydratase (DHAD) mutant-V497F | | Descriptor: | ACETATE ION, Dihydroxy-acid dehydratase, chloroplastic, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Dihydroxyacid dehydratase (DHAD) mutant-V497F
To Be Published
|
|
8IKZ
 
 | | The mutant structure of DHAD | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The mutant structure of DHAD
To Be Published
|
|
5XJ6
 
 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the glycerol 3-phosphate form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION, ... | | Authors: | Li, Z, Tang, Y, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
5XJ8
 
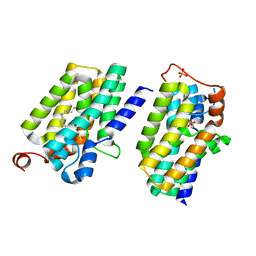 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the lysphosphatidic acid form | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION | | Authors: | Li, Z, Tang, Y, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
5ZE4
 
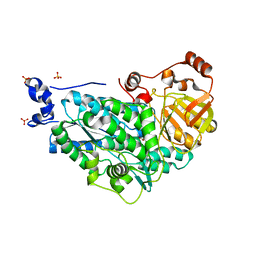 | | The structure of holo- structure of DHAD complex with [2Fe-2S] cluster | | Descriptor: | ACETATE ION, Dihydroxy-acid dehydratase, chloroplastic, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y, Gan, J, Wu, L. | | Deposit date: | 2018-02-26 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action.
Nature, 559, 2018
|
|
5ZI6
 
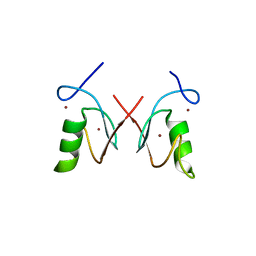 | | The RING domain structure of MEX-3C | | Descriptor: | RNA-binding E3 ubiquitin-protein ligase MEX3C, ZINC ION | | Authors: | Moududee, S.A, Tang, Y. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of hMEX-3C Ring finger domain as an E3 ubiquitin ligase
Protein Sci., 27, 2018
|
|
6J3N
 
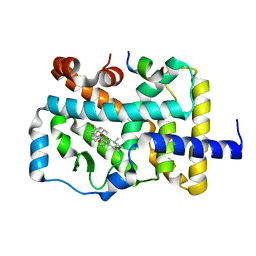 | | RORgammat LBD complexed with Ursonic Acid and SRC2.2 | | Descriptor: | (5beta)-3-oxours-12-en-28-oic acid, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Nuclear receptor ROR-gamma | | Authors: | Liu, Z.H, Huang, J, Tang, Y. | | Deposit date: | 2019-01-05 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of human RORgammat LBD with SCR2.2 at 1.99 Angstroms resolution
To Be Published
|
|
7EAX
 
 | | Crystal complex of p53-V272M and antimony ion | | Descriptor: | ANTIMONY (III) ION, Cellular tumor antigen p53, ZINC ION | | Authors: | Lu, M, Tang, Y. | | Deposit date: | 2021-03-08 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Repurposing antiparasitic antimonials to noncovalently rescue temperature-sensitive p53 mutations.
Cell Rep, 39, 2022
|
|
7CZ0
 
 | | Crystal structure of a thermostable green fluorescent protein (TGP) with a synthetic nanobody (Sb92) | | Descriptor: | ACETATE ION, CACODYLATE ION, CACODYLIC ACID, ... | | Authors: | Cai, H, Yao, H, Li, T, Hutter, C, Tang, Y, Li, Y, Seeger, M, Li, D. | | Deposit date: | 2020-09-06 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | An improved fluorescent protein tag and its nanobodies for membrane protein expression, stability assay, and purification
To Be Published
|
|
7ENQ
 
 | | Crystal structure of human NAMPT in complex with compound NAT | | Descriptor: | 2-(2-~{tert}-butylphenoxy)-~{N}-(4-hydroxyphenyl)ethanamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Wang, G, Wu, C, Liu, M, Yao, H, Li, C, Wang, L, Tang, Y. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.204966 Å) | | Cite: | Discovery of small-molecule activators of nicotinamide phosphoribosyltransferase (NAMPT) and their preclinical neuroprotective activity.
Cell Res., 32, 2022
|
|
7E3M
 
 | | RORgamma LBD complexed with Panaxatriol and SRC2.2 | | Descriptor: | (3R,5R,6S,8R,9R,10R,12R,13R,14R,17S)-4,4,8,10,14-pentamethyl-17-[(2R)-2,6,6-trimethyloxan-2-yl]-2,3,5,6,7,9,11,12,13,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthrene-3,6,12-triol, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Nuclear receptor ROR-gamma | | Authors: | Liu, Z.H, Huang, J, Lu, W.Q, Tang, Y, Wu, Z.R. | | Deposit date: | 2021-02-09 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human RORgammat LBD with SRC2.2 at 2.80 Angstroms resolution
To Be Published
|
|
7EGV
 
 | | Acetolactate Synthase from Trichoderma harzianum with inhibitor harzianic acid | | Descriptor: | (2S)-3-methyl-2-[[(2S,4R)-1-methyl-4-[(2E,4E)-octa-2,4-dienoyl]-3,5-bis(oxidanylidene)pyrrolidin-2-yl]methyl]-2-oxidanyl-butanoic acid, 1,2-ETHANEDIOL, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Zang, X, Xie, L, Chen, M, Tang, Y, Zhou, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Harzianic Acid from Trichoderma afroharzianum Is a Natural Product Inhibitor of Acetohydroxyacid Synthase.
J.Am.Chem.Soc., 2021
|
|
7EHE
 
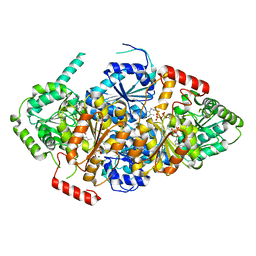 | | Acetolactate Synthase from Trichoderma harzianum | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Acetolactate synthase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zang, X, Tang, Y, Zhou, J. | | Deposit date: | 2021-03-29 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Harzianic Acid from Trichoderma afroharzianum Is a Natural Product Inhibitor of Acetohydroxyacid Synthase.
J.Am.Chem.Soc., 2021
|
|
5YM0
 
 | | The crystal structure of DHAD | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, SULFATE ION | | Authors: | Zang, X, Huang, W.X, Cheng, R, Wu, L, Zhou, J.H, Tang, Y, Yan, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | The crystal structure of DHAD
To Be Published
|
|
