7F8J
 
 | | Cryo-EM structure of human pannexin-1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8O
 
 | | Cryo-EM structure of the C-terminal deletion mutant of human PANX1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
2KWC
 
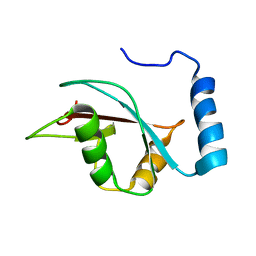 | | The NMR structure of the autophagy-related protein Atg8 | | Descriptor: | Autophagy-related protein 8 | | Authors: | Kumeta, H, Watanabe, M, Nakatogawa, H, Yamaguchi, M, Ogura, K, Adachi, W, Fujioka, Y, Noda, N.N, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-04-05 | | Release date: | 2010-05-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the autophagy-related protein Atg8
J.Biomol.Nmr, 47, 2010
|
|
1UAW
 
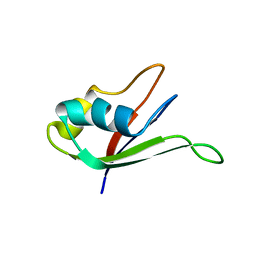 | | Solution structure of the N-terminal RNA-binding domain of mouse Musashi1 | | Descriptor: | mouse-musashi-1 | | Authors: | Miyanoiri, Y, Kobayashi, H, Watanabe, M, Ikeda, T, Nagata, T, Okano, H, Uesugi, S, Katahira, M. | | Deposit date: | 2003-03-24 | | Release date: | 2004-03-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics
J.Biol.Chem., 278, 2003
|
|
5XL0
 
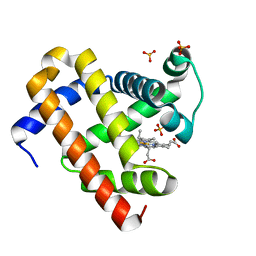 | | met-aquo form of sperm whale myoglobin reconstituted with 7-PF, a heme possesseing CF3 group as side chain | | Descriptor: | Myoglobin, SULFATE ION, fluorinated heme | | Authors: | Kanai, Y, Harada, A, Shibata, T, Nishimura, R, Namiki, K, Watanabe, M, Nakamura, S, Yumoto, F, Senda, T, Suzuki, A, Neya, S, Yamamoto, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Characterization of Heme Orientational Disorder in a Myoglobin Reconstituted with a Trifluoromethyl-Group-Substituted Heme Cofactor
Biochemistry, 56, 2017
|
|
5YOT
 
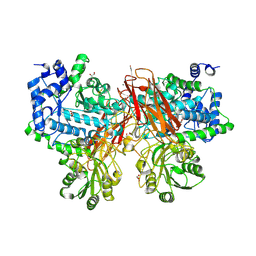 | | Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Matsuzawa, T, Watanabe, M, Nakamichi, Y, Yaoi, K. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure and substrate recognition mechanism of Aspergillus oryzae isoprimeverose-producing enzyme.
J.Struct.Biol., 205, 2019
|
|
1IT8
 
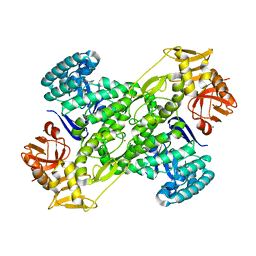 | | Crystal structure of archaeosine tRNA-guanine transglycosylase from Pyrococcus horikoshii complexed with archaeosine precursor, preQ0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-01-11 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
1IQ8
 
 | | Crystal Structure of archaeosine tRNA-guanine transglycosylase from Pyrococcus horikoshii | | Descriptor: | ARCHAEOSINE TRNA-GUANINE TRANSGLYCOSYLASE, MAGNESIUM ION, ZINC ION | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-07-09 | | Release date: | 2002-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
1IT7
 
 | | Crystal structure of archaeosine tRNA-guanine transglycosylase complexed with guanine | | Descriptor: | Archaeosine tRNA-guanine transglycosylase, GUANINE, MAGNESIUM ION, ... | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-01-11 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
3WQ8
 
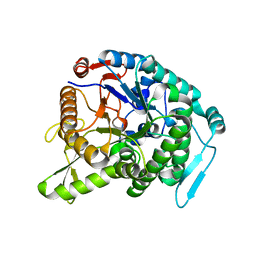 | | Monomer structure of hyperthermophilic beta-glucosidase mutant forming a dodecameric structure in the crystal form | | Descriptor: | Beta-glucosidase | | Authors: | Nakabayashi, M, Kataoka, M, Watanabe, M, Ishikawa, K. | | Deposit date: | 2014-01-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Monomer structure of a hyperthermophilic beta-glucosidase mutant forming a dodecameric structure in the crystal form.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
8JOR
 
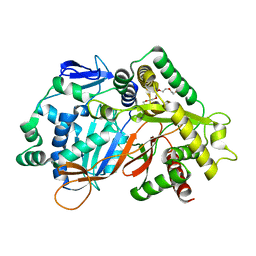 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type A crystal | | Descriptor: | Acyltransferase, PENTAETHYLENE GLYCOL | | Authors: | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
8JOS
 
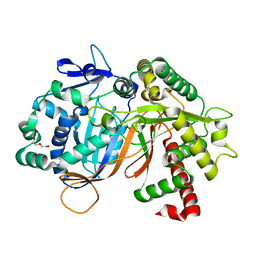 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type B crystal | | Descriptor: | Acyltransferase, CHLORIDE ION, TRIETHYLENE GLYCOL | | Authors: | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
2CZW
 
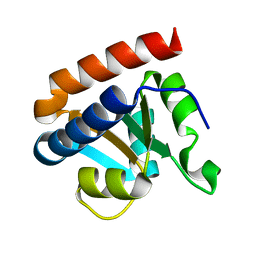 | | Crystal structure analysis of protein component Ph1496p of P.horikoshii ribonuclease P | | Descriptor: | 50S ribosomal protein L7Ae | | Authors: | Fukuhara, H, Kifusa, M, Watanabe, M, Terada, A, Honda, T, Numata, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3
Biochem.Biophys.Res.Commun., 343, 2006
|
|
2YU0
 
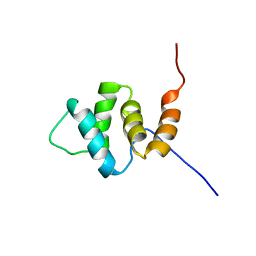 | | Solution structures of the PAAD_DAPIN domain of mus musculus interferon-activatable protein 205 | | Descriptor: | Interferon-activable protein 205 | | Authors: | Sato, M, Tochio, N, Koshiba, S, Watanabe, M, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the PAAD_DAPIN domain of mus musculus interferon-activatable protein 205
To be Published
|
|
5YQS
 
 | | Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yaoi, K, Matsuzawa, T, Watanabe, M, Nakamichi, Y. | | Deposit date: | 2017-11-07 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and substrate recognition mechanism of Aspergillus oryzae isoprimeverose-producing enzyme.
J.Struct.Biol., 205, 2019
|
|
5ZN6
 
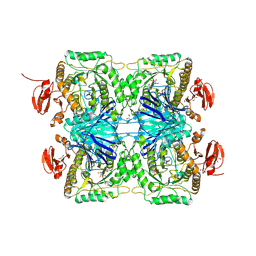 | |
7EAP
 
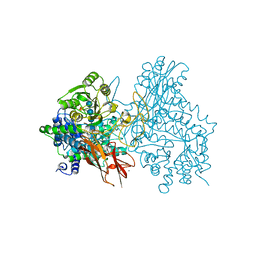 | | Crystal structure of IpeA-XXXG complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Matsuzawa, T, Watanabe, M, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis for the catalytic mechanism of the glycoside hydrolase family 3 isoprimeverose-producing oligoxyloglucan hydrolase from Aspergillus oryzae.
Febs Lett., 596, 2022
|
|
5ZN7
 
 | |
2DVY
 
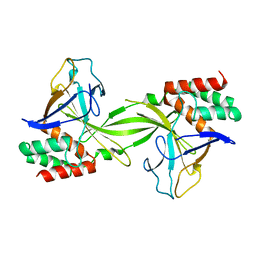 | | Crystal structure of restriction endonucleases PabI | | Descriptor: | Restriction endonuclease PabI | | Authors: | Miyazono, K, Watanabe, M, Kamo, M, Sawasaki, T, Nagata, K, Endo, Y, Tanokura, M, Kobayashi, I. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel protein fold discovered in the PabI family of restriction enzymes
Nucleic Acids Res., 35, 2007
|
|
6JOW
 
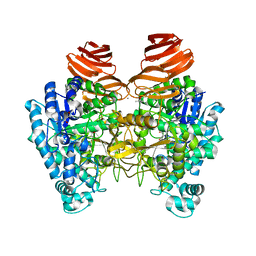 | |
6KFF
 
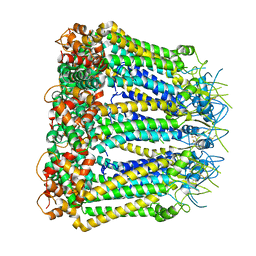 | | Undocked INX-6 hemichannel in a nanodisc | | Descriptor: | Innexin-6 | | Authors: | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2019-07-07 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
6KFH
 
 | | Undocked hemichannel of an N-terminal deletion mutant of INX-6 in a nanodisc | | Descriptor: | Innexin-6 | | Authors: | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2019-07-07 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
6KRN
 
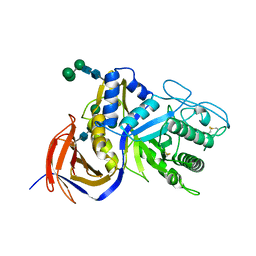 | | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris in complex with aldotriuronic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, Mating factor alpha,GH30 Xylanase B, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Substrate recognition by a bifunctional GH30-7 xylanase B from Talaromyces cellulolyticus.
Febs Open Bio, 10, 2020
|
|
6KRL
 
 | | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Substrate recognition by a bifunctional GH30-7 xylanase B from Talaromyces cellulolyticus.
Febs Open Bio, 10, 2020
|
|
