7KZ3
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, SODIUM ION | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZ5
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, ... | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZD
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the reduced internal aldimine and with bound Glutarate | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 1,2-ETHANEDIOL, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, ... | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
2YSW
 
 | | Crystal Structure of the 3-dehydroquinate dehydratase from Aquifex aeolicus VF5 | | Descriptor: | 3-dehydroquinate dehydratase | | Authors: | Tanaka, T, Kumarevel, T.S, Ebihara, A, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-04 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the 3-dehydroquinate dehydratase from Aquifex aeolicus VF5
To be Published
|
|
5Z4B
 
 | | GB1 structure determination in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Protein LG | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
7CNL
 
 | | Crystal structure of TEAD3 in complex with VT105 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, N-[(1S)-1-pyridin-2-ylethyl]-8-[4-(trifluoromethyl)phenyl]quinoline-3-carboxamide, ... | | Authors: | Tang, T.T, Konradi, A.W. | | Deposit date: | 2020-08-01 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Small Molecule Inhibitors of TEAD Auto-palmitoylation Selectively Inhibit Proliferation and Tumor Growth of NF2 -deficient Mesothelioma.
Mol.Cancer Ther., 20, 2021
|
|
1KOT
 
 | |
7CLJ
 
 | | Crystal structure of Thermoplasmatales archaeon heliorhodopsin E108D mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, RETINAL, SULFATE ION, ... | | Authors: | Tanaka, T, Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-07-21 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for unique color tuning mechanism in heliorhodopsin.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7DDZ
 
 | | The Crystal Structure of Human Neuropeptide Y Y2 Receptor with JNJ-31020028 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Human Neuropeptide Y Y2 Receptor fusion protein, ~{N}-[4-[4-[(1~{S})-2-(diethylamino)-2-oxidanylidene-1-phenyl-ethyl]piperazin-1-yl]-3-fluoranyl-phenyl]-2-pyridin-3-yl-benzamide | | Authors: | Tang, T, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2020-10-30 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for ligand recognition of the neuropeptide Y Y 2 receptor.
Nat Commun, 12, 2021
|
|
1BXD
 
 | | NMR STRUCTURE OF THE HISTIDINE KINASE DOMAIN OF THE E. COLI OSMOSENSOR ENVZ | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN (OSMOLARITY SENSOR PROTEIN (ENVZ)) | | Authors: | Tanaka, T, Saha, S.K, Tomomori, C, Ishima, R, Liu, D, Tong, K.I, Park, H, Dutta, R, Qin, L, Swindells, M.B, Yamazaki, T, Ono, A.M, Kainosho, M, Inouye, M, Ikura, M. | | Deposit date: | 1998-10-02 | | Release date: | 1999-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ.
Nature, 396, 1998
|
|
5DMQ
 
 | |
2ZWI
 
 | | Crystal structure of alpha/beta-Galactoside alpha-2,3-Sialyltransferase from a Luminous Marine Bacterium, Photobacterium phosphoreum | | Descriptor: | Alpha-/beta-galactoside alpha-2,3-sialyltransferase, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Iwatani, T, Okino, N, Sakakura, M, Kajiwara, H, Ichikawa, M, Takakura, Y, Kimura, M, Ito, M, Yamamoto, T, Kakuta, Y. | | Deposit date: | 2008-12-05 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of alpha/beta-galactoside alpha2,3-sialyltransferase from a luminous marine bacterium, Photobacterium phosphoreum
Febs Lett., 583, 2009
|
|
6IR7
 
 | | Green fluorescent protein variant GFPuv with the modification to 6-hydroxynorleucine at the C-terminus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-HYDROXY-L-NORLEUCINE, Green fluorescent protein, ... | | Authors: | Nakatani, T, Yasui, N, Yamashita, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.277 Å) | | Cite: | Specific modification at the C-terminal lysine residue of the green fluorescent protein variant, GFPuv, expressed in Escherichia coli.
Sci Rep, 9, 2019
|
|
6IR6
 
 | | Green fluorescent protein variant GFPuv with the native lysine residue at the C-terminus | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Nakatani, T, Yasui, N, Yamashita, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Specific modification at the C-terminal lysine residue of the green fluorescent protein variant, GFPuv, expressed in Escherichia coli.
Sci Rep, 9, 2019
|
|
7CCN
 
 | | The binding structure of a lanthanide binding tag (LBT3) with lutetium ion (Lu3+) | | Descriptor: | LBT3, LUTETIUM (III) ION | | Authors: | Hatanaka, T, Kikkawa, N, Matsugami, A, Hosokawa, Y, Hayashi, F, Ishida, N. | | Deposit date: | 2020-06-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The origins of binding specificity of a lanthanide ion binding peptide.
Sci Rep, 10, 2020
|
|
7CCO
 
 | | The binding structure of a lanthanide binding tag (LBT3) with lanthanum ion (La3+) | | Descriptor: | LANTHANUM (III) ION, LBT3 | | Authors: | Hatanaka, T, Kikkawa, N, Matsugami, A, Hosokawa, Y, Hayashi, F, Ishida, N. | | Deposit date: | 2020-06-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The origins of binding specificity of a lanthanide ion binding peptide.
Sci Rep, 10, 2020
|
|
4W4Q
 
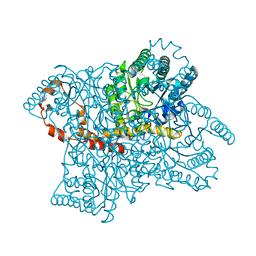 | | Glucose isomerase structure determined by serial femtosecond crystallography at SACLA | | Descriptor: | CALCIUM ION, Xylose isomerase | | Authors: | Nango, E, Tanaka, T, Sugahara, M, Suzuki, M, Iwata, S. | | Deposit date: | 2014-08-15 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat.Methods, 12, 2015
|
|
6THE
 
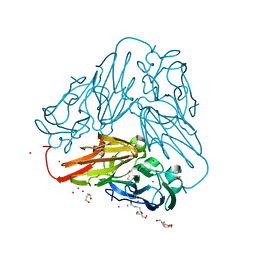 | | Crystal structure of core domain of four-domain heme-cupredoxin-Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
6LPK
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 293 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6TBR
 
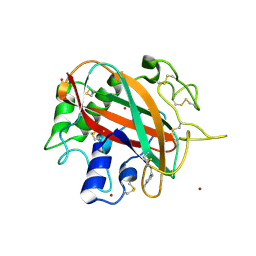 | | Glycosylated AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae in P1 space group | | Descriptor: | AoAA13, ZINC ION | | Authors: | Frandsen, K.E.H, Muderspach, S.J, Tandrup, T, Poulsen, J.C.N, Lo Leggio, L. | | Deposit date: | 2019-11-04 | | Release date: | 2020-03-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Further structural studies of the lytic polysaccharide monooxygenase AoAA13 belonging to the starch-active AA13 family
Amylase, 3(1), 2019
|
|
6TFO
 
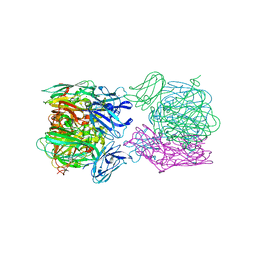 | | Crystal structure of as isolated three-domain copper-containing nitrite reductase from Hyphomicrobium denitrificans strain 1NES1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of substrate- and product-bound forms of a multi-domain copper nitrite reductase shed light on the role of domain tethering in protein complexes.
Iucrj, 7, 2020
|
|
3GFJ
 
 | |
5MM0
 
 | | Dolichyl phosphate mannose synthase in complex with GDP-mannose and Mn2+ (donor complex) | | Descriptor: | CHLORIDE ION, Dolichol monophosphate mannose synthase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Gandini, R, Reichenbach, T, Tan, T.C, Divne, C. | | Deposit date: | 2016-12-08 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for dolichylphosphate mannose biosynthesis.
Nat Commun, 8, 2017
|
|
5MM1
 
 | | Dolichyl phosphate mannose synthase in complex with GDP and dolichyl phosphate mannose | | Descriptor: | Dolichol monophosphate mannose synthase, GUANOSINE-5'-DIPHOSPHATE, dolichyl phosphate mannose | | Authors: | Gandini, R, Reichenbach, T, Tan, T.C, Divne, C. | | Deposit date: | 2016-12-08 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for dolichylphosphate mannose biosynthesis.
Nat Commun, 8, 2017
|
|
5MLZ
 
 | | Dolichyl phosphate mannose synthase in complex with GDP and Mg2+ | | Descriptor: | CHLORIDE ION, Dolichol monophosphate mannose synthase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gandini, R, Reichenbach, T, Tan, T.C, Divne, C. | | Deposit date: | 2016-12-08 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for dolichylphosphate mannose biosynthesis.
Nat Commun, 8, 2017
|
|
