2EE6
 
 | | Solution structure of the 21th filamin domain from human Filamin-B | | Descriptor: | Filamin-B | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-15 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 21th filamin domain from human Filamin-B
To be Published
|
|
2EA5
 
 | | Solution Structure of the RING domain of the human Cell growth regulator with RING finger domain 1 protein | | Descriptor: | Cell growth regulator with RING finger domain protein 1, ZINC ION | | Authors: | Miyamoto, K, Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-30 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the RING domain of the human Cell growth regulator with RING finger domain 1 protein
To be Published
|
|
2EE9
 
 | | Solution structure of the 16th filamin domain from human Filamin-B | | Descriptor: | Filamin-B | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-15 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 16th filamin domain from human Filamin-B
To be Published
|
|
2EE7
 
 | | Solution structure of the CH domain from human Sperm flagellar protein 1 | | Descriptor: | Sperm flagellar protein 1 | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-15 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain from human Sperm flagellar protein 1
To be Published
|
|
2YT5
 
 | | Solution structure of the PHD domain of Metal-response element-binding transcription factor 2 | | Descriptor: | Metal-response element-binding transcription factor 2, ZINC ION | | Authors: | Masuda, K, Muto, Y, Isono, K, Watanabe, S, Harada, T, Kigawa, T, Koseki, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PHD domain of Metal-response element-binding transcription factor 2
To be Published
|
|
2RR6
 
 | | Solution structure of the leucine rich repeat of human acidic leucine-rich nuclear phosphoprotein 32 family member B | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Authors: | Tochio, N, Umehara, T, Tsuda, K, Koshiba, S, Harada, T, Watanabe, S, Tanaka, A, Kigawa, T, Yokoyama, S. | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain.
J.Mol.Biol., 401, 2010
|
|
3VX3
 
 | | Crystal structure of [NiFe] hydrogenase maturation protein HypB from Thermococcus kodakarensis KOD1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ... | | Authors: | Sasaki, D, Watanabe, S, Miki, K. | | Deposit date: | 2012-09-09 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Structure of a Novel Archaeal HypB for [NiFe] Hydrogenase Maturation
J.Mol.Biol., 425, 2013
|
|
3WJQ
 
 | | Crystal structure of the HypE CN form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BENZAMIDINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tominaga, T, Watanabe, S, Miki, K. | | Deposit date: | 2013-10-14 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WJP
 
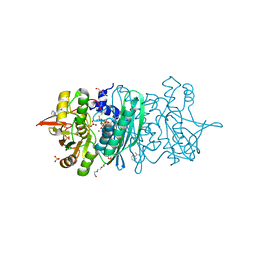 | | Crystal structure of the HypE CA form | | Descriptor: | BENZAMIDINE, GLYCEROL, Hydrogenase expression/formation protein HypE, ... | | Authors: | Tominaga, T, Watanabe, S, Miki, K. | | Deposit date: | 2013-10-14 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.533 Å) | | Cite: | Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WJR
 
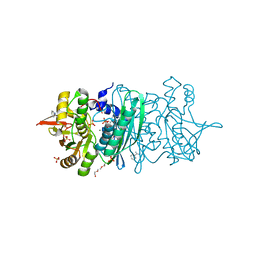 | | crystal structure of HypE in complex with a nucleotide | | Descriptor: | ADENOSINE MONOPHOSPHATE, BENZAMIDINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tominaga, T, Watanabe, S, Miki, K. | | Deposit date: | 2013-10-14 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7BYU
 
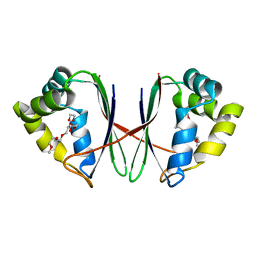 | | Crystal structure of Acidovorax avenae L-fucose mutarotase (apo form) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, L-fucose mutarotase | | Authors: | Watanabe, Y, Fukui, Y, Watanabe, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Functional and structural characterization of a novel L-fucose mutarotase involved in non-phosphorylative pathway of L-fucose metabolism.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
7C0E
 
 | |
7C0C
 
 | |
7CGR
 
 | |
7CGQ
 
 | |
6GEV
 
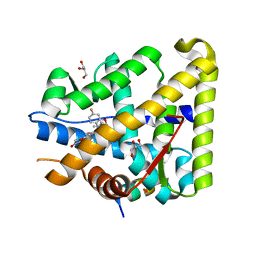 | | Mineralocorticoid receptor in complex with (s)-13 | | Descriptor: | 6-[[(3~{S})-7-fluoranyl-3-(2-methylpropyl)-2,3-dihydro-1,4-benzoxazin-4-yl]carbonyl]-4~{H}-1,4-benzoxazin-3-one, GLYCEROL, Mineralocorticoid receptor, ... | | Authors: | Edman, K, Aagaard, A, Tangefjord, S. | | Deposit date: | 2018-04-27 | | Release date: | 2019-01-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of Mineralocorticoid Receptor Modulators with Low Impact on Electrolyte Homeostasis but Maintained Organ Protection.
J.Med.Chem., 62, 2019
|
|
6GG8
 
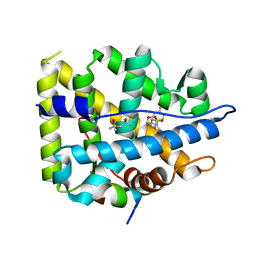 | | Mineralocorticoid receptor in complex with (s)-13 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Mineralocorticoid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Edman, K, Aagaard, A, Tangefjord, S. | | Deposit date: | 2018-05-03 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Mineralocorticoid Receptor Modulators with Low Impact on Electrolyte Homeostasis but Maintained Organ Protection.
J.Med.Chem., 62, 2019
|
|
7LTO
 
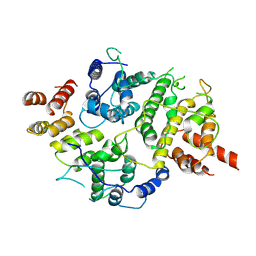 | | Nse5-6 complex | | Descriptor: | Non-structural maintenance of chromosome element 5, Ubiquitin-like protein SMT3,DNA repair protein KRE29 chimera | | Authors: | Yu, Y, Li, S.B, Zheng, S, Tangy, S, Koyi, C, Wan, B.B, Kung, H.H, Andrej, S, Alex, K, Patel, D.J, Zhao, X.L. | | Deposit date: | 2021-02-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Integrative analysis reveals unique structural and functional features of the Smc5/6 complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CNQ
 
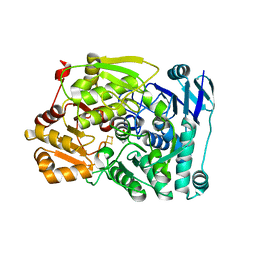 | | Crystal structure of Agrobacterium tumefaciens aconitase X (holo-form) | | Descriptor: | (2~{S},3~{R})-3-oxidanylpyrrolidine-2-carboxylic acid, FE2/S2 (INORGANIC) CLUSTER, cis-3-hydroxy-L-proline dehydratase | | Authors: | Murase, Y, Watanabe, Y, Watanabe, S. | | Deposit date: | 2020-08-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
7CNR
 
 | |
7CNP
 
 | |
7CNS
 
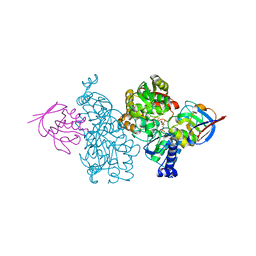 | | Crystal structure of Thermococcus kodakaraensis aconitase X (holo-form) | | Descriptor: | (3R)-3-HYDROXY-3-METHYL-5-(PHOSPHONOOXY)PENTANOIC ACID, DUF521 domain-containing protein, FE3-S4 CLUSTER, ... | | Authors: | Murase, Y, Watanabe, Y, Watanabe, S. | | Deposit date: | 2020-08-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
7D2R
 
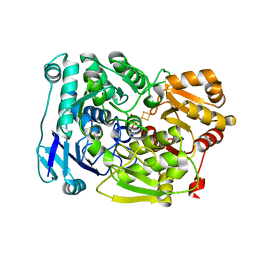 | | Crystal structure of Agrobacterium tumefaciens aconitase X mutant - S449C/C510V | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, SODIUM ION, ... | | Authors: | Murase, Y, Watanabe, Y, Watanabe, S. | | Deposit date: | 2020-09-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
6UX6
 
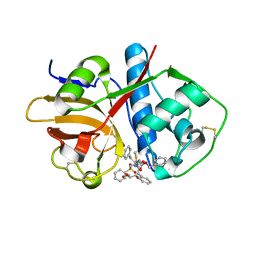 | | Cruzain covalently bound by a vinylsulfone compound | | Descriptor: | Cruzipain, GLYCEROL, Nalpha-[(benzyloxy)carbonyl]-N-[(2S)-1-phenyl-4-(phenylsulfonyl)butan-2-yl]-L-phenylalaninamide | | Authors: | Zhai, X, Tang, S, Chenna, B.C, Meek, T.D, Sacchettini, J.C. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Cruzain covalently bound by a vinylsulfone compound
To Be Published
|
|
2EYY
 
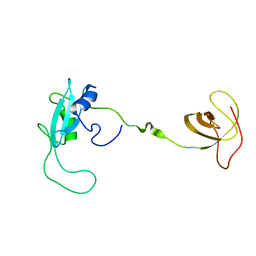 | | CT10-Regulated Kinase isoform I | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
