8QC0
 
 | |
9CLR
 
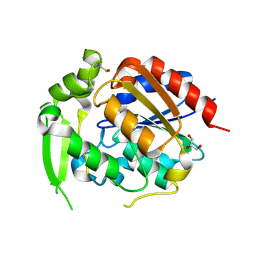 | | Hydroxynitrile Lyase from Hevea brasiliensis with Seventy-one Mutations | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tan, P, Meixner, E.L, Nguyen, A, Kazlaukas, R.J, Pierce, C.T, Evans, R.L, Shi, K, Aihara, H. | | Deposit date: | 2024-07-12 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Hydroxynitrile Lyase from Hevea brasiliensis with Seventy-one Mutations
To Be Published
|
|
9EMW
 
 | |
9EMX
 
 | |
8PQQ
 
 | |
8PQR
 
 | | Nucleoside 2'deoxyribosyltransferase from Chroococcidiopsis thermalis PCC 7203 WT bound to DAD_Immucillin-H | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Nucleoside 2-deoxyribosyltransferase | | Authors: | Tang, P, Harding, C.J, Czekster, C.M. | | Deposit date: | 2023-07-11 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.586 Å) | | Cite: | Snapshots of the Reaction Coordinate of a Thermophilic 2'-Deoxyribonucleoside/ribonucleoside Transferase.
Acs Catalysis, 14, 2024
|
|
8PQT
 
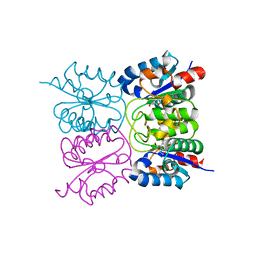 | |
8PQS
 
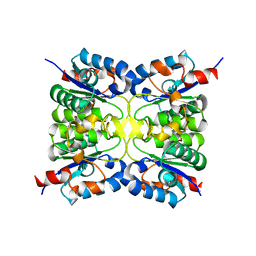 | |
8PQP
 
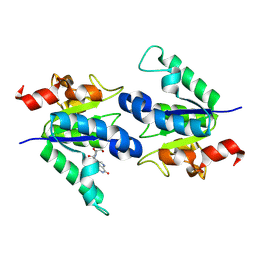 | |
3D4F
 
 | | SHV-1 beta-lactamase complex with LN1-255 | | Descriptor: | (3R)-4-{[(3,4-dihydroxyphenyl)acetyl]oxy}-N-(2-formylindolizin-3-yl)-3-sulfino-D-valine, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Pattanaik, P, Bethel, C.R, Hujer, A.M, Bonomo, R.A, Buynak, J.D, van den Akker, F. | | Deposit date: | 2008-05-14 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Strategic Design of an Effective {beta}-Lactamase Inhibitor: LN-1-255, A 6-ALKYLIDENE-2'-SUBSTITUTED PENICILLIN SULFONE
J.Biol.Chem., 284, 2009
|
|
8RH3
 
 | | Nucleoside 2'deoxyribosyltransferase from Chroococcidiopsis thermalis PCC 7203 WT bound to Gemcitabine | | Descriptor: | (2~{R},3~{R})-4,4-bis(fluoranyl)-2-(hydroxymethyl)oxolan-3-ol, Nucleoside 2-deoxyribosyltransferase | | Authors: | Tang, P, Harding, C.J, Czekster, C.M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Snapshots of the Reaction Coordinate of a Thermophilic 2'-Deoxyribonucleoside/ribonucleoside Transferase.
Acs Catalysis, 14, 2024
|
|
7TDZ
 
 | |
5I8U
 
 | | Crystal Structure of the RV1700 (MT ADPRASE) E142Q mutant | | Descriptor: | ADP-ribose pyrophosphatase, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Thirawatananond, P, Kang, L.-W, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and mutational studies of the adenosine diphosphate ribose hydrolase from Mycobacterium tuberculosis.
J. Bioenerg. Biomembr., 48, 2016
|
|
5W6Z
 
 | | Crystal structure of the H24W mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
5VY2
 
 | | Crystal structure of the F36A mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-05-24 | | Release date: | 2018-11-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
5W6X
 
 | |
5WJI
 
 | | Crystal structure of the F61S mutant of HsNUDT16 | | Descriptor: | ACETIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-07-23 | | Release date: | 2018-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
8WIO
 
 | | Durio zibethinus trypsin inhibitor DzTI-10 | | Descriptor: | 21 kDa seed protein-like, SULFATE ION | | Authors: | Deentanya, P, Wangkanont, K. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Kunitz-type trypsin inhibitor from durian (Durio zibethinus) employs a distinct loop for trypsin inhibition.
Protein Sci., 33, 2024
|
|
8WIN
 
 | | Durio zibethinus trypsin inhibitor DzTI-12 | | Descriptor: | 21 kDa seed protein-like | | Authors: | Deentanya, P, Wangkanont, K. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kunitz-type trypsin inhibitor from durian (Durio zibethinus) employs a distinct loop for trypsin inhibition.
Protein Sci., 33, 2024
|
|
8WFO
 
 | | Durio zibethinus trypsin inhibitor DzTI-4 | | Descriptor: | 21 kDa seed protein-like, ACETATE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Deentanya, P, Wangkanont, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Kunitz-type trypsin inhibitor from durian (Durio zibethinus) employs a distinct loop for trypsin inhibition.
Protein Sci., 33, 2024
|
|
8WI1
 
 | | Durio zibethinus trypsin inhibitor DzTI-8 | | Descriptor: | 21 kDa seed protein-like | | Authors: | Deentanya, P, Wangkanont, K. | | Deposit date: | 2023-09-24 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kunitz-type trypsin inhibitor from durian (Durio zibethinus) employs a distinct loop for trypsin inhibition.
Protein Sci., 33, 2024
|
|
8WK1
 
 | |
8WKB
 
 | | Durio zibethinus trypsin inhibitor DzTI-7 | | Descriptor: | 21 kDa seed protein-like, CALCIUM ION, GLYCEROL | | Authors: | Deentanya, P, Wangkanont, K. | | Deposit date: | 2023-09-27 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Kunitz-type trypsin inhibitor from durian (Durio zibethinus) employs a distinct loop for trypsin inhibition.
Protein Sci., 33, 2024
|
|
8WQ6
 
 | |
4GD8
 
 | | SHV-1 beta-lactamase in complex with penam sulfone SA3-53 | | Descriptor: | (2S,3R)-4-(2-aminoethylcarbamoyloxy)-2-[(2-methanoylindolizin-3-yl)amino]-3-methyl-3-sulfino-butanoic acid, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Pattanaik, P, van den Akker, F. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of SHV-1 beta-lactamase with penem and penam sulfone inhibitors that form cyclic intermediates stabilized by carbonyl conjugation
Plos One, 7, 2012
|
|
